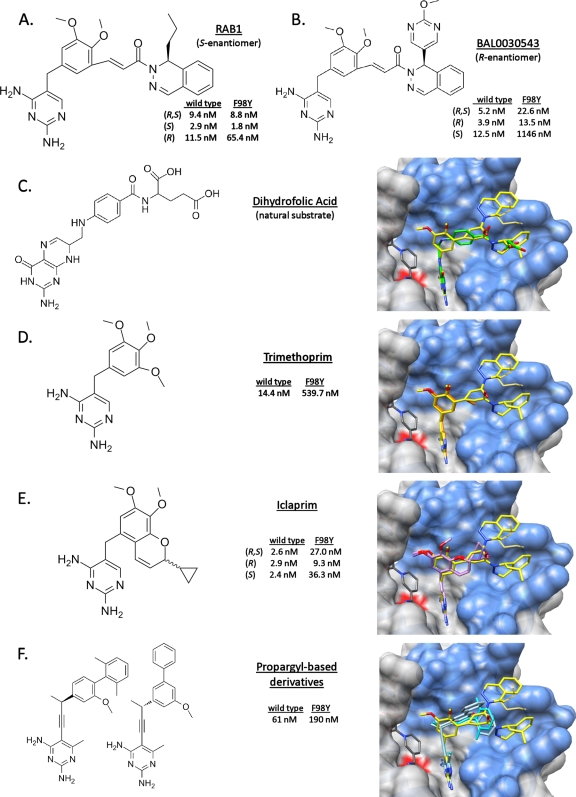
FIG. 2.
RAB1 occupies a similar space as the natural substrate and other inhibitors; in addition it utilizes a unique surface cavity. The IC50 of RAB1 is not affected by the F98Y mutation to the same extent as other inhibitors. RAB1 is depicted as yellow in the binding site (blue), residue 98 is marked with a red surface, and the listed small molecules are superimposed. (A) RAB1 with IC50 data from the current study. (B) BAL0030543, a related compound under development against S. aureus, with IC50 data from reference 7. (C) Dihydrofolic acid, with coordinates obtained from PDB ID 3FRD (26) (D) Trimethoprim, with IC50 data from the current study and the coordinates obtained from PDB ID 2W9G with NADPH (22). (E) Iclaprim, with IC50 and coordinates obtained from PDB IDs 3FYV and 3FYW (27). (F) Propargyl-based inhibitors, with IC50 data and coordinates obtained from PDB ID 3F0B (19) and from PDB ID 3F0Q (K. M. Frey et al., unpublished; no IC50 data are publicly available).