Table 1. X-ray data-collection and refinement statistics for Ub2 .
Values in parentheses are for the last shell.
| X-ray source | ESRF ID29 |
| Wavelength (Å) | 0.97625 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 58.7, b = 78.7, c = 93.1, α = γ = 90, β = 97.9 |
| Mosaicity (°) | 0.30 |
| Images | 180 |
| Oscillation angle (°) | 1.0 |
| Resolution (Å) | 39.90–1.60 (1.69–1.60) |
| Unique reflections | 54118 (7792) |
| Completeness (%) | 97.9 (96.8) |
| Multiplicity | 3.8 (3.8) |
| 〈I〉/〈σ(I)〉 | 16.1 (3.2) |
| Rmerge† | 0.057 (0.432) |
| Solvent content (%) | 41 |
| No. of reflections in Rfree set (5%) | 2738 |
| Rwork | 0.183 |
| Rfree | 0.229 |
| FOM | 0.851 |
| R.m.s. deviations from ideal values‡ | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.5 |
| Torsion angles (°) | 6.1 |
| Protein atoms | 3962 |
| Water atoms | 360 |
| Ligand atoms (1 ethylene glycol, 3 sulfate ions) | 19 |
| Disordered residues (not modelled) | Chain B, 76; chains D, F, 74, 75, 76§ |
| Average B factors (Å2) | |
| Protein main chain | 19 |
| Protein side chain | 21 |
| Water | 32 |
| Ethylene glycol | 28 |
| Sulfate ions | 58 |
| Ramachandran outliers¶ | 1 [Gln62 in chain D] |
| Estimated coordinate error†† (Å) | 0.18 |
| PDB code | 3m3j |
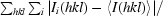
 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value for all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value for all i measurements.
Ideal values as reported in Engh & Huber (2001 ▶).
These residues correspond to the C-termini of proximal ubiquitin moieties.
Residues for which the backbone torsion angles are outside the core region of the Ramachandran plot (Kleywegt & Jones, 1996 ▶).
Coordinate error estimated from a Luzzati plot (R/R free versus resolution) as reported by SFCHECK (Vaguine et al., 1999 ▶).
