Table 1. Crystal data and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Binary Dpo4Mg2+ | Dpo4dGTPMg2+ | Dpo4dCTPMg2+ | |
|---|---|---|---|
| Type of lesion | 1,N 2--G | 8-oxoG | 8-oxoG |
| Crystal data and data collection | |||
| X-ray source | APS | APS | APS |
| Beamline | SER-CAT 22-BM | DND-CAT 5-ID | SER-CAT 22-ID |
| Detector | MAR CCD 300 | MAR CCD 225 | MAR CCD 300 |
| Wavelength () | 1.00 | 1.00 | 1.00 |
| Temperature (K) | 110 | 110 | 110 |
| No. of crystals | 1 | 1 | 1 |
| Space group | P21212 | P21212 | P21 |
| Unit-cell parameters | |||
| a () | 93.72 | 94.44 | 59.75 |
| b () | 102.12 | 103.90 | 100.42 |
| c () | 53.00 | 52.70 | 105.87 |
| () | 90 | 90 | 90 |
| () | 90 | 90 | 96.07 |
| () | 90 | 90 | 90 |
| Resolution range () | 50.02.2 (2.282.2) | 29.132.5 (2.662.5) | 49.52.6 (2.762.6) |
| No. of measurements | 331946 | 106973 | 195670 |
| No. of unique reflections | 26245 (4241) | 18457 (3005) | 37629 (5685) |
| Redundancy | 12.6 (10.4) | 5.8 (5.3) | 5.2 (3.8) |
| Completeness (%) | 99.3 (99.3) | 99.6 (99.9) | 98.1 (90.1) |
| R merge † | 6.2 (48.2) | 6.4 (48.6) | 6.8 (49.4) |
| I/(I) | 36.3 (6.2) | 21.3 (4.2) | 14.8 (3.7) |
| Solvent content (%) | 48.1 | 50.1 | 59.6 |
| Refinement | |||
| Model composition (asymmetric unit) | |||
| No. of amino-acid residues | 341 | 341 | 341/342 |
| No. of water molecules | 247 | 192 | 305 |
| No. of Mg2+ ions | 0 | 4 | 3/3 |
| No. of template nucleotides | 16 | 17 | 17/17 |
| No. of primer nucleotides | 14 | 13 | 13/13 |
| No. of dGTP | 1 | ||
| No. of dCTP | 1/1 | ||
| R ‡ (%) | 22.1 | 22.0 | 22.3 |
| R free § (%) | 25.0 | 25.9 | 26.8 |
| Estimated coordinate error () | |||
| Luzzati plot | 0.30 | 0.32 | 0.34 |
| Luzzati plot (cross-validation) | 0.35 | 0.40 | 0.42 |
| A plot | 0.28 | 0.35 | 0.42 |
| A plot (cross-validation) | 0.30 | 0.43 | 0.49 |
| Temperature factors | |||
| Wilson plot (2) | 41.3 | 57.4 | 47.0 |
| Mean isotropic (2) | 40.7 | 44.1 | 48.3 |
| Root-mean-square deviation in temperature factors | |||
| Bonded main-chain atoms (2) | 1.4 | 1.3 | 1.3 |
| Bonded side-chain atoms (2) | 2.0 | 1.9 | 1.9 |
| Root-mean-square deviation from ideal values | |||
| Bond lengths () | 0.007 | 0.007 | 0.007 |
| Bond angles () | 1.5 | 1.4 | 1.3 |
| Dihedral angles () | 21.3 | 21.6 | 22.0 |
| Improper angles () | 1.1 | 1.6 | 1.0 |
R
merge = 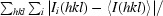
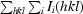 , where the outer sum (hkl) is taken over the unique reflections.
, where the outer sum (hkl) is taken over the unique reflections.
R = 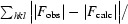
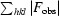 , where |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively.
, where |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively.
R free is the same as R but calculated for a set of reflections (5% of the total) omitted from the refinement process.
