In this study, the glucansucrase from the dental caries pathogen S. mutans was purified and crystallized by the hanging-drop vapour-diffusion method using ammonium sulfate as a precipitant.
Keywords: glucansucrase, dental caries, Streptococcus mutans
Abstract
Glucansucrases encoded by Streptococcus mutans play essential roles in the synthesis of sticky dental plaques. Based on amino-acid sequence similarity, glucansucrases are classified as members of glycoside hydrolase family 70 (GH 70). Data on the crystal structure of GH 70 glucansucrases have yet to be reported. Here, the GH 70 glucansucrase GTF-SI from S. mutans was overexpressed in Escherichia coli strain BL21 (DE3), purified to homogeneity and crystallized using the hanging-drop vapour-diffusion method. Orthorhombic GTF-SI crystals belonging to space group P21212 were obtained. A diffraction data set was collected to 2.1 Å resolution.
1. Introduction
Streptococcus mutans is closely associated with the pathogenesis of dental caries (Hamada & Slade, 1980 ▶). The mechanism of dental caries involves the initial formation of a biofilm, or dental plaque, comprised of a sticky glucose polymer (glucan) that is produced by S. mutans in which oral bacteria, food debris and salivary components can become trapped. Acids derived from the fermentation of dietary carbohydrates such as sucrose, fructose and glucose lead to the demineralization of the adjacent tooth surface and ultimately to dental caries. Sticky glucan plays an essential role in the aetiology and pathogenesis of dental caries (Yamashita et al., 1992 ▶). Dental plaques also cause co-pathologies such as infection, periodontal disease and halitosis (Liu et al., 2009 ▶). Sticky glucan is synthesized from sucrose by glucansucrases (GSases) encoded by S. mutans. Therefore, these enzymes are attractive drug targets for the development of effective treatments for dental caries (Newbrun et al., 1983 ▶). Several inhibitors of GSases from S. mutans have been identified that can prevent dental caries (Yanagida et al., 2000 ▶; Koo et al., 2002 ▶). Detailed structural information on GSases would be instrumental in the development of more effective treatments; however, to date crystallographic structural analyses of S. mutans GSases have yet to be reported.
GSase is a glycosyl transferase (GTF) and a member of glycoside hydrolase family 70 (GH 70; http://afmb.cnrs-mrs.fr/CAZY/). GSase is a large enzyme, with an average molecular weight of 160 kDa. GSases contain an N-terminal variable region, a conserved catalytic domain of approximately 900 amino acids and a C-terminal domain of approximately 500 amino acids composed of a series of homologous directly repeating units that are responsible for glucan binding (Monchois et al., 1999 ▶). The regions of GSase that are essential for enzyme catalysis have been precisely identified (Monchois et al., 1999 ▶; van Hijum et al., 2006 ▶), but the detailed mechanisms of regioselectivity and stereospecificity remain unclear.
In the oral cavity, sticky glucan synthesis by S. mutans is mediated by three extracellular GSases: GTF-SI, GTF-I and GTF-S (Tamesada et al., 2004 ▶). GTF-SI and GTF-I are mainly involved in the synthesis of insoluble glucan containing α(1–3) glycosidic linkages, while GTF-S is primarily involved in the synthesis of soluble α(1–6)-linked glucan (Monchois et al., 1999 ▶; van Hijum et al., 2006 ▶). Together, these three enzymes function cooperatively to produce insoluble sticky glucan.
Here, we present a crystallization report for the GSase GTF-SI from the dental caries pathogen S. mutans, including a description of the expression, purification, crystallization and preliminary X-ray analysis of GTF-SI.
2. Materials and methods
2.1. Gene cloning
S. mutans strain MT8148 was obtained from RIKEN BioResource Center, Japan. Genomic DNA was extracted and purified using a DNeasy kit (Qiagen). The gene sequence corresponding to the catalytic domain of GTF-SI (amino-acid residues 244–1163) was amplified by PCR and then subcloned into pCold I (Takara) using the restriction enzymes NdeI and XhoI to generate pCold-GTF-SI. In this way, the N-terminal variable region and C-terminal glucan-binding domain were removed from the protein. The sequences of the gene-specific primers used for PCR were as follows: 5′-CATATGGTCAAAAATATCAGAAAAGTGAACGGT-3′ and 5′-CTCGAGACCATCAAATACCAATCCAGTTACA-3′.
2.2. Protein expression and purification.
Recombinant GTF-SI was produced in Escherichia coli BL21 (DE3) as follows. Briefly, E. coli harbouring pCold-GTF-SI were inoculated into 1 l LB medium containing 100 µg ml−1 ampillicin. Cultures were grown at 310 K to an OD600 of 0.5. The expression of recombinant GTF-SI was induced by cold treatment and the cells were then incubated for an additional 24 h at 288 K. The enzyme was purified using TALON metal-affinity resin (Clontech). Purified GTF-SI was dialyzed against 10 mM MES buffer pH 6.3 and then concentrated to 30 mg ml−1 with a Centricon YM-50 filter (Millipore). The purity of the protein was assessed by SDS–PAGE.
2.3. Crystallization
Crystallization screening was carried out in VDX plates (Hampton Research) at 293 and 277 K by the hanging-drop vapour-diffusion method using Crystal Screen and Crystal Screen 2 (Hampton Research). Crystals were obtained under conditions that contained ammonium sulfate at 293 K. Further screening was performed using Detergent Screen and Additive Screen (Hampton Research). Crystal growth was carried out in hanging drops under optimized conditions using the microseeding technique at 293 K by mixing 1.5 µl protein solution (30 mg ml−1) with 1.5 µl reservoir solution containing 0.1 M MES buffer pH 6.5, 1.6 M ammonium sulfate and 0.2% ANAPOE-C10E9.
2.4. Data collection and processing
A crystal of dimensions 0.2 × 0.1 × 0.1 mm was mounted in a cryoloop, soaked in cryoprotectant consisting of 30% glycerol for 60 s and then flash-frozen in liquid nitrogen. Diffraction data from the GTF-SI crystal were collected on beamline NE3A of the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan). Data-collection and data-processing statistics are presented in Table 1 ▶. The data sets were processed and scaled using DENZO and SCALEPACK from the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
Table 1. Data-collection statistics for GTF-SI.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 50.00–2.10 (2.15–2.10) |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 293.88, b = 215.42, c = 218.95 |
| Unique reflections | 753230 (54729) |
| Completeness | 100.0 (99.9) |
| Redundancy | 7.0 (5.9) |
| Rmerge† (%) | 7.7 (44.1) |
| 〈I/σ(I)〉 | 22.6 (3.6) |
| Mosaicity | 0.369 |
| Unit-cell volume (Å3) | 1.39 × 107 |
R
merge = 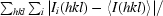
 , where 〈I(hkl)〉 is the mean intensity of equivalent reflections.
, where 〈I(hkl)〉 is the mean intensity of equivalent reflections.
3. Results and discussion
Alignment analysis of the primary sequences of GH 70 GSases and glycoside hydrolase family 13 α-amylases revealed evidence of a circular permutation between these two families in the secondary-structural elements of the TIM barrel of the catalytic domain (MacGregor et al., 1996 ▶). It is believed that these two families of enzymes share a mechanism of transglycosylation at the single catalytic site (Monchois et al., 1999 ▶; van Hijum et al., 2006 ▶). Here, we subcloned the catalytic domain of GTF-SI from S. mutans strain MT8148. The GTF-SI catalytic region contained 844 amino acids and had a calculated molecular mass of 94.4 kDa. The amino-acid sequence of the GTF-SI catalytic domain had 58% and 90% identity to the corresponding domains from GTF-S and GTF-I, respectively. In the recombinant protein, the N-terminal variable region of GTF-SI was replaced by a histidine tag and the C-terminal glucan-binding domain was removed. Diffraction was not improved by removal of the histidine tag (data not shown). Purified recombinant GTF-SI migrated as a single band with an apparent molecular weight of 95 kDa on SDS–PAGE (Fig. 1 ▶). Approximately 20 mg purified GTF-SI was obtained from a 1 l culture. After three months of crystallization trials, spherical crystals were obtained under condition No. 23 of Hampton Crystal Screen 2, which consists of 0.1 M MES buffer pH 6.5, 1.6 M ammonium sulfate and 10% dioxane. No other conditions produced crystals. A manual two-dimensional grid screen around the hit condition and screening for additive compounds were also performed. The crystal quality was greatly improved by the use of ANAPOE-C10E9 as a detergent. However, the crystals grew in cluster form. Good-quality plate-shaped monocrystals appeared after one week using the microseeding technique (Fig. 2 ▶). The diffraction pattern is shown in Fig. 3 ▶. A single crystal with dimensions of 0.2 × 0.1 × 0.1 mm was used for diffraction experiments and data were collected to 2.1 Å resolution under cryogenic conditions. A 180° data set was collected with an oscillation angle of 0.3° and an exposure time of 2 s per frame. The diffraction data were indexed in the orthorhombic space group P21212, with unit-cell parameters a = 293.88, b = 215.42, c = 218.95 Å. Calculation of the Matthews coefficient based on a molecular weight of 94.35 kDa yielded 12–20 molecules per asymmetric unit (V M = 3.1–1.8 Å3 Da−1 with 60–33% solvent content, respectively). The data-collection statistics are summarized in Table 1 ▶. Owing to low sequence similarity and structural discrepancies, the GTF-SI crystal data could not be phased by the molecular-replacement method using homologous proteins for which tertiary structures are available in the Protein Data Bank (such as Bacillus licheniformis α-amylase, which has 23% identity). Structure determination using selenomethionine-derivatized GTF-SI from S. mutans is currently in progress.
Figure 1.
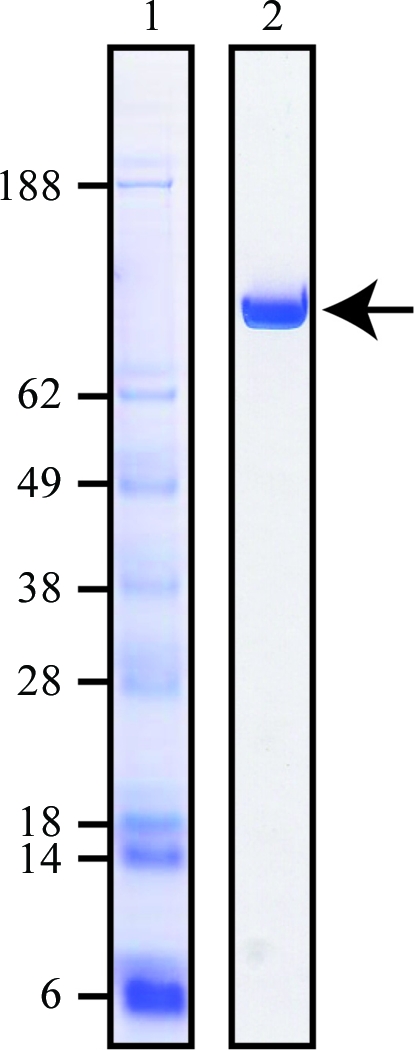
SDS–PAGE analysis of purified recombinant GTF-SI. GTF-SI was analyzed by SDS–PAGE and visualized by Commassie Brilliant Blue staining. Lane 1, protein molecular-weight markers (kDa); lane 2, purified recombinant GTF-SI catalytic domain.
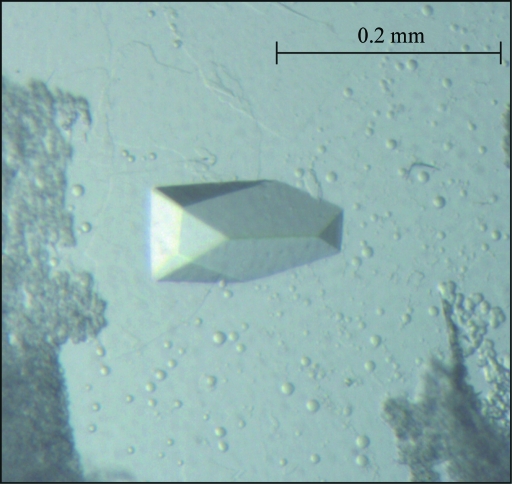
Figure 2.
Image of a crystal of S. mutans GTF-SI. The plate-shaped monocrystal appeared after one week using the hanging-drop vapour-diffusion method and a microseeding technique. The largest dimension of the crystal was approximately 0.2 mm.
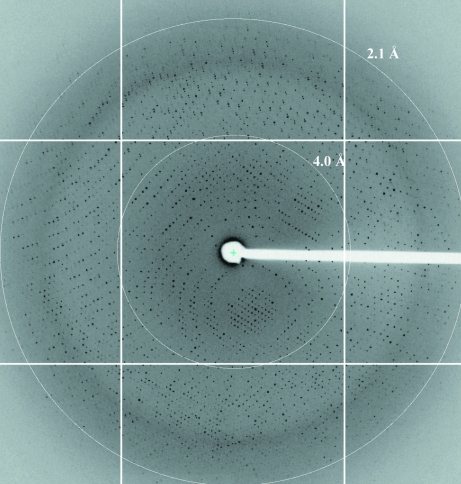
Figure 3.
Diffraction pattern of a GTF-SI crystal. The crystal was grown under optimum conditions and then flash-frozen in glycerol-containing cryoprotectant. The exposure time was 2 s, with an oscillation angle of 0.3°. The pattern displayed a maximum resolution of 2.1 Å and space group P21212.
Acknowledgments
This study was supported in part by the ERATO Iwata Human Receptor Crystallography Project (JST; to S. Iwata), a Grant-in-Aid for Young Scientists (B) (to S. Ito) and Grants-in-Aid for Scientific Research (B) (to TK and TS) in Japan. The X-ray diffraction data collection was approved by the Photon Factory Advisory Committee (Proposal No. 2010G169).
References
- Hamada, S. & Slade, H. D. (1980). Microbiol. Rev.44, 331–384. [DOI] [PMC free article] [PubMed]
- Hijum, S. A. van, Kralj, S., Ozimek, L. K., Dijkhuizen, L. & van Geel-Schutten, I. G. (2006). Microbiol. Mol. Biol. Rev.70, 157–176. [DOI] [PMC free article] [PubMed]
- Koo, H., Rosalen, P. L., Cury, J. A., Park, Y. K. & Bowen, W. H. (2002). Antimicrob. Agents Chemother.46, 1302–1309. [DOI] [PMC free article] [PubMed]
- Liu, P.-F., Zhu, W.-H. & Huang, C.-M. (2009). Curr. Drug Metab.10, 90–94. [DOI] [PMC free article] [PubMed]
- MacGregor, E. A., Jespersen, H. M. & Svensson, B. (1996). FEBS Lett.378, 263–266. [DOI] [PubMed]
- Monchois, V., Willemot, R. M. & Monsan, P. (1999). FEMS Microbiol. Rev.23, 131–151. [DOI] [PubMed]
- Newbrun, E., Hoover, C. I. & Walker, G. J. (1983). Arch. Oral Biol.28, 531–536. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tamesada, M., Kawabata, S., Fujiwara, T. & Hamada, S. (2004). J. Dent. Res.83, 874–879. [DOI] [PubMed]
- Yamashita, Y., Bowen, W. H. & Kuramitsu, H. K. (1992). Infect. Immun.60, 1618–1624. [DOI] [PMC free article] [PubMed]
- Yanagida, A., Kanda, T., Tanabe, M., Matsudaira, F. & Cordeiro, J. G. O. (2000). J. Agric. Food Chem.48, 5666–5671. [DOI] [PubMed]