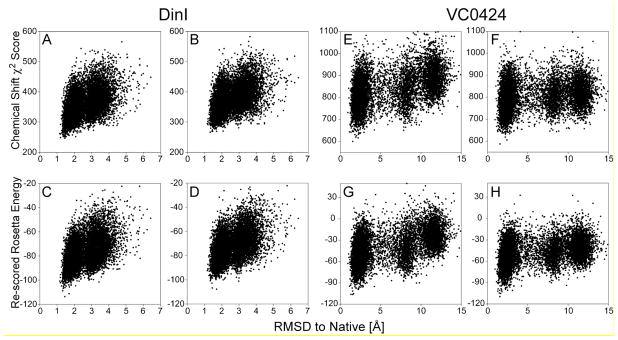
Figure 3.
CS-Rosetta model selection using either SPARTA+ or SPARTA chemical shift predictions. For proteins DinI (A-D; PDB entry 1GHH (Ramirez et al. 2000)) and VC0424 (E-H; PDB entry 1NXI (Ramelot et al. 2003)), 10,000 structures each were generated by a standard CS-Rosetta protocol, using a protein structural database with chemical shifts added by SPARTA+. For each CS-Rosetta model, the totalχ2 error function between the experimental chemical shifts and values predicted by SPARTA+ (A,E) or SPARTA (B,F) are plotted against the Cα coordinate rmsd relative to the experimental PDB structure. The re-scored Rosetta energy, calculated by adding the scaled SPARTA+ (C,G) or SPARTA (D,H) chemical shift χ2 score to the raw Rosetta energy, is also plotted and used to select the final models (Table S3).