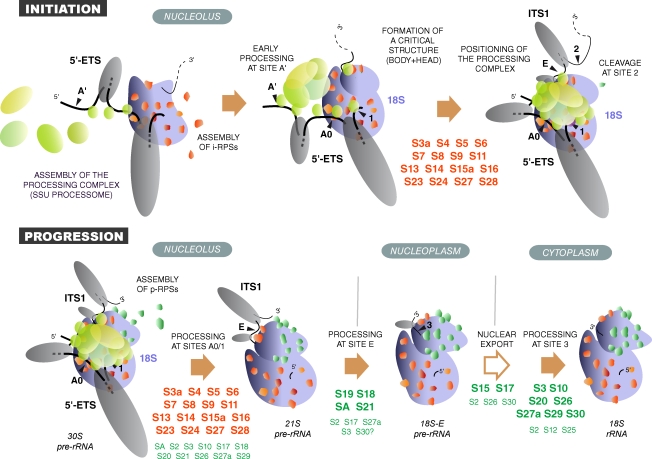
Figure 9.
Model of RPS protein activity in ribosome biogenesis. i-RPS proteins (red) assemble with the nascent pre-rRNA, most likely cotranscriptionally (Chooi and Leiby, 1981), together with early preribosomal factors (blue), in particular UTP proteins and sno-RNPs, like U3 and U17. i-RPS proteins participate in the folding of the 18S rRNA domain, whereas large secondary structures form in the transcribed spacers (Renalier et al., 1989; Michot and Bachellerie, 1991), potentially involving the binding of preribosomal factors. This allows formation of an assembly intermediate in which the processing machinery is correctly positioned, thus initiating cleavage of the 5′-ETS. p-RPSs (green) are mostly involved in downstream steps, although they may be necessary for coordination of processing at sites A0 and 1. The pre-40S particles are released from the nucleolus after cleavage at site E and are exported to the cytoplasm where the final processing step occurs. The RPS proteins required at each processing step are indicated.