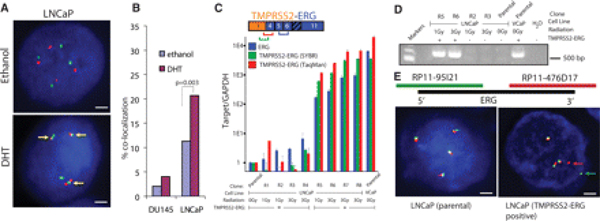
Figure 1.
(A) FISH based evaluation of induced proximity between TMPRSS2 (green) and ERG (red) on stimulation with 100 nM DHT in LNCaP cells. Colocalization is indicated by yellow arrows. Scale bar indicate 2 µm. (B) Induced proximity between TMPRSS2 and ERG in DU145 and LNCaP cells is quantified and represented as the percentage of nuclei exhibiting colocalization signals. GAPDH, glyceraldehydes-3-phosphate dehydrogenase. (C) SYBR Green (Moleculr Probes, Eugene, Orgeon) and TaqMan (applied Biosystems, Foster City, California) quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of the TMPRSS2-ERG fusion transcript. (D) Gel based RT-PCR analysis with primers spanning the first exon of TMPRSS2 and sixth exon of ERG for representative clones. (E) FISH analysis of the ERG locus. Split signals representing ERG rearrangement are highlighted by arrows.