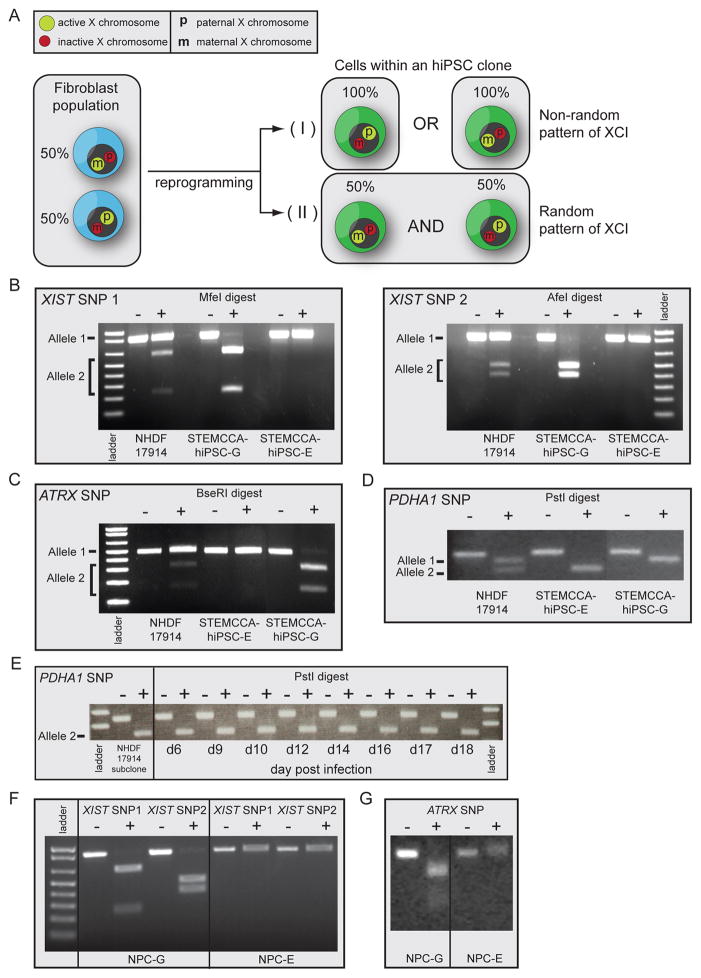
Figure 4. Non-random XCI in female hiPSC lines.
(A) Schematic representation of random XCI in fibroblasts and potential XCI states in female hiPSC lines.
(B) Analysis of allele-specific expression of XIST using restriction enzyme-sensitive SNPs. RNA from NHDF17914 fibroblasts and STEMCCA-hiPSC lines E and G (passages 8–9) derived from this fibroblast population was reverse transcribed and two regions of XIST surrounding different SNPs were amplified by PCR and analyzed with and without the respective restriction digest as indicated. SNPs were chosen such that the PCR product derived from the two X alleles are differentially cleaved as indicated. Note that fibroblasts express both XIST alleles, while hiPSC line G exclusively expresses one and line E the other allele.
(C) As in (B) except for allele-specific expression of ATRX.
(D) As in (B) except for allele-specific expression of PDHA1.
(E) A subclone of NHDF17914 fibroblasts obtained upon delivery of a lentiviral shRNA targeting p53 was reprogrammed with the STEMCCA approach and allele-specific expression of PDHA1 determined at different times of the reprogramming process using the same assay as in (D). This fibroblast subclone only expresses allele 2 and does not reactivate allele 1 during reprogramming.
(F) As in (B) but, allele-specific expression of XIST was analyzed upon differentiation of STEMCCA hiPSC lines E and G into neural precursors (NPCs), indicating that non-random XCI is preserved upon differentiation.
(G) As in (F) except for allele-specific expression of ATRX in hiPSC-derived NPCs. See Fig S4 for additional information.