Figure 1.
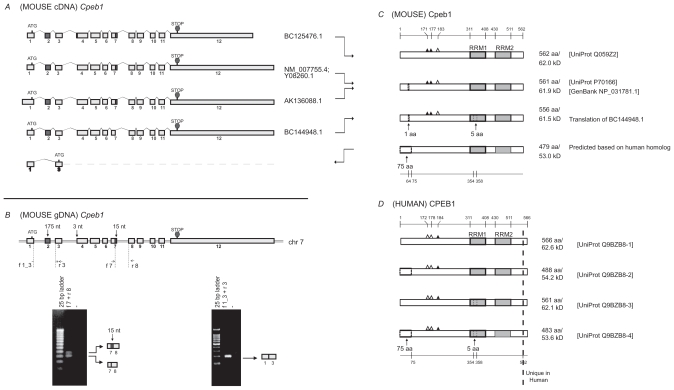
Analysis of Cpeb1. A) Transcripts of mouse Cpeb1. Only non-redundant full-length UniGene sequences were used for this analysis, with their accession numbers listed on the right. ATG and STOP indicate the presence of translational initiation and termination sites, respectively. The different lengths of the first and last exons likely represent the presence of variable 5′ and 3′ UTRs, respectively. The alternative splices of the first 3-nt of exon 4 and the last 15-nt of exon 7 (highlighted in grey) would generate different protein products. A novel isoform with deletion of exon 2 (also highlighted in grey) is predicted based on published sequences of the human protein. The deletion of exon 2 leads to the use of an alternative translational start codon in exon 3. B) Expression of Cpeb1 transcripts in adult mouse retina. The locations of the primers for RT-PCR are aligned to the diagram of Cpeb1 genomic DNA, in which boxes represent exons and double lines represent introns. Photographs of DNA gels demonstrate the expression of multiple Cpeb1 transcripts in the retina including those with and without the 15-nt of exon 7 (left), and without exon 2 (right). The identity of each band was confirmed by nucleotide sequencing. C) Isoforms of mouse Cpeb1 proteins. Two isoforms were extracted from the UniProt database (Q059Z2, P70166). The computational translation of cDNA BC144948.1 yields a third isoform. A fourth isoform is predicted based on two human CPEB1 homologs (Q9BZB8-2, Q9BZB8-4). RNA recognition motifs (RRMs) are indicated with grey boxes. Triangles represent phosphorylation sites experimentally confirmed (solid)21 or predicted (open) based on cross-ortholog comparisons. A 1-aa deletion, a 5-aa deletion, and a 75-aa N-terminal truncation are each indicated with dashed line boxes. The locations of functional motifs are shown as numbered amino acid sites at the top of the diagram, and those of the alternative spliced regions at the bottom, as might be seen in the longest isoform. D) Isoforms of human CPEB1 proteins. Four isoforms are extracted from the UniProt database. The RRMs, the phosphorylation sites, and the 5-aa deletion are all present in human CPEB1 at the same locations as seen in mouse. Two isoforms have 75-aa N terminal truncations (Q9BZB8-2, Q9BZB8-4). This led to our predication of the existence of a similar isoform in mouse. Human CPEB1 has an additional 4-aa at the C-terminus than mouse Cpeb1 (shown to the right of the dashed line). Numeric annotations refer to the longest isoform.