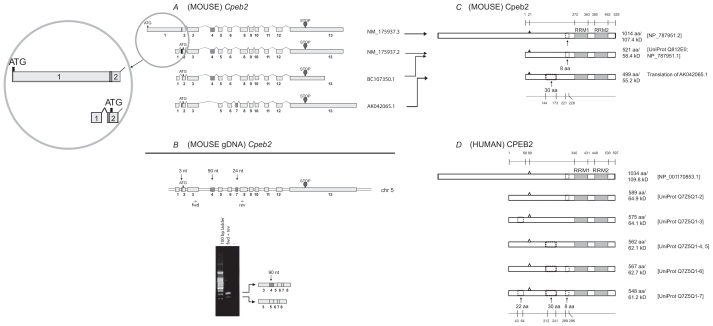
Figure 2.
Analysis of Cpeb2. A) Transcripts of mouse Cpeb2. Three previously established non-redundant full-length UniGene sequences and a newly updated sequence (the top one) are used for the analysis, with their accession numbers listed on the right of the figure. ATG and STOP indicate the presence of translational initiation and termination sites, respectively. The different lengths of the first and the last exons likely represent the presence of variable 5′ UTR and 3′ UTR, respectively. The alternative splice of the first 3-nt of exon 2 also result in a different 5′ UTR. The alternative splices of exon 4 (90-nt) or exon 7 (24-nt) would generate different protein products. Insert: the previous version and the new version of NM_175937 sequences are both legitimate. In the new version, the first exon was extended towards both ends and “fused” with exon 2. This would lead to a much longer cDNA and a longer N terminus in the protein. B) Expression of Cpeb2 transcripts in adult mouse retina. The locations of the primers for RT-PCR are aligned to the diagram of Cpeb2 genomic DNA, in which boxes represent exons and double lines represent introns. The photograph of DNA Gel demonstrates the expression of two Cpeb2 transcripts in the retina with and without exon4. The identity of each band was confirmed with nucleotide sequencing. C) Isoforms of mouse Cpeb2 proteins. The previously established isoform (Q812E0; NP_787951.1) has an 8-aa deletion. The newly updated isoform (NP_787951.2) has an 8-aa deletion as well as a much longer N-terminus, which is due to the use of a longer exon 1 in cDNA NM_175937.3. The computational translation of cDNA AK042065.1 generates an additional isoform which has a 30-aa deletion. RRMs are indicated with gray boxes. Triangles represent phosphorylation sites experimentally confirmed.22 A 30-aa deletion and an 8-aa deletion are indicated in dashed line boxes. The locations of functional motifs are shown as numbered amino acid sites at the top of the diagram, and those of the alternative spliced regions at the bottom, as might be seen in a conceptual isoform without any deletion. D) Isoforms of human CPEB2 proteins. Five isoforms are extracted from the UniProt database. The RRMs, the phosphorylation sites (open triangles, predicted based on cross-ortholog comparisons), and the two deletions are all present in human CPEB2 at similar locations. The recently updated, unusually long isoform of human CPEB2 (NP_001170853.1) was aligned to its closest isoform. The first 68-aa in the previously established human CPEB2 isoforms was thought to be a region that was unique for human, but now aligns to a region in the extra-long isoform of mouse Cpeb2.