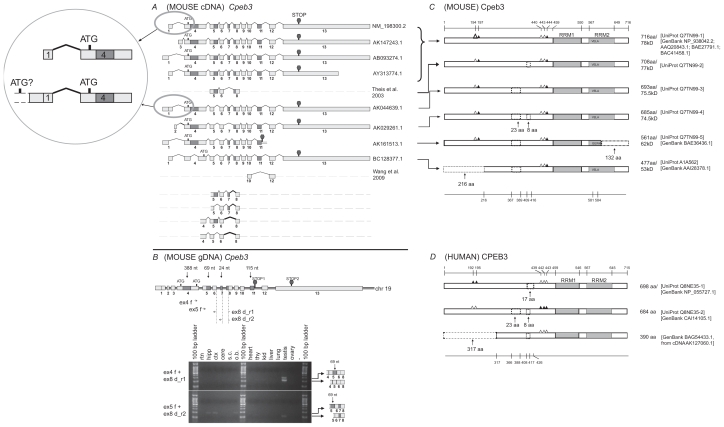
Figure 3.
Analysis of Cpeb3. A) Transcripts of mouse Cpeb3. Eight non-redundant full-length UniGene sequences were used for the analysis, with the accession numbers listed on the right. ATG and STOP indicate the presence of translational initiation and termination sites, respectively. Partial sequences of two transcripts were derived from previous publications.5,6 The different lengths of the first and the last exons likely represent different 5′ UTR and 3′ UTR, respectively. The alternative splices of exon 2 and 3 would also result in different 5′ UTRs. The intra-exon skipping of exon 4 (388-nt), the partial skipping of exon 5 (69-nt), and the deletion of exon 7 (24-nt) or exon 11 (115-nt) would generate different protein products. Exon 11 extension (AK161513.1) would lead to an early translational termination and an altered 4-aa at the C-terminus. Four additional alternatively splice variants, all with a 27-nt skipping at the beginning of exon 8, were identified in this study. Insert: The comparison between two cDNAs that use different 5′ UTRs. The upstream elongation of exon 1 in AK044639.1, with additional alternative splice(s), may lead to the use of an alternative translation start codon, which would generate an extralong isoform of Cpeb3 protein with an extended N-terminus. B) Expression of Cpeb3 transcripts in adult mouse retina. The locations of the primers for RT-PCR are aligned to the diagram of Cpeb3 genomic DNA, in which boxes represent exons and double lines represent introns. Photographs of DNA gel demonstrate the expression of four Cpeb3 transcripts in the retina without the 27-nt in exon 8, and with or without the 24-nt (exon 7) and the 69-nt (exon 5). The identity of each band was confirmed with nucleotide sequencing. Many of the other alternatively spliced regions have been confirmed in previous publications5,6 in great details. Tissue abbreviation: rtn—retina, hipp—hippocampus, ctx—cortex, cere—cerebellum, s.c.—spinal cord, o.b.—olfactory bulb, thy—thymus, kid—kidney. C) Isoforms of mouse Cpeb3 proteins. Six isoforms are extracted from the UniProt database. RRMs were indicated with gray boxes. Triangles represent phosphorylation sites experimentally confirmed21,24,25 (solid) or predicted (open) according to cross-paralog comparisons, respectively. A 216-aa N-terminal truncation, two internal deletions of 23-aa motif and 8-aa motif, and a 132-aa C terminal truncation with altered C terminus were indicated in dashed lines. The C-terminal truncation (Q7TN99-5) removes the majority of the second RRM and alters the last four amino acids from VELA to GEWK. The locations of functional motifs are shown as numbered amino acid sites at the top of the diagram, and those of the alternative spliced regions at the bottom, as might be seen in the longest isoform. D) Isoforms of human CPEB3 proteins. Three isoforms are extracted from Uni- Prot and NCBI databases. The RRMs, the phosphorylation sites, and the 23-aa and 8-aa deletions are all present in human CPEB3 at similar locations. Numeric annotations refer to a conceptual isoform without any deletion.