Abstract
Genomics and human genetics are scientifically fundamental and commercially valuable. These fields grew to prominence in an era of growth in government and nonprofit research funding, and of even greater growth of privately funded research and development in biotechnology and pharmaceuticals. Patents on DNA technologies are a central feature of this story, illustrating how patent law adapts---and sometimes fails to adapt---to emerging genomic technologies. In instrumentation and for therapeutic proteins, patents have largely played their traditional role of inducing investment in engineering and product development, including expensive postdiscovery clinical research to prove safety and efficacy. Patents on methods and DNA sequences relevant to clinical genetic testing show less evidence of benefits and more evidence of problems and impediments, largely attributable to university exclusive licensing practices. Whole-genome sequencing will confront uncertainty about infringing granted patents but jurisprudence trends away from upholding the broadest and potentially most troublesome patent claims.
Keywords: intellectual property, licensing, DNA patents, gene patents, open source, law
BACKGROUND
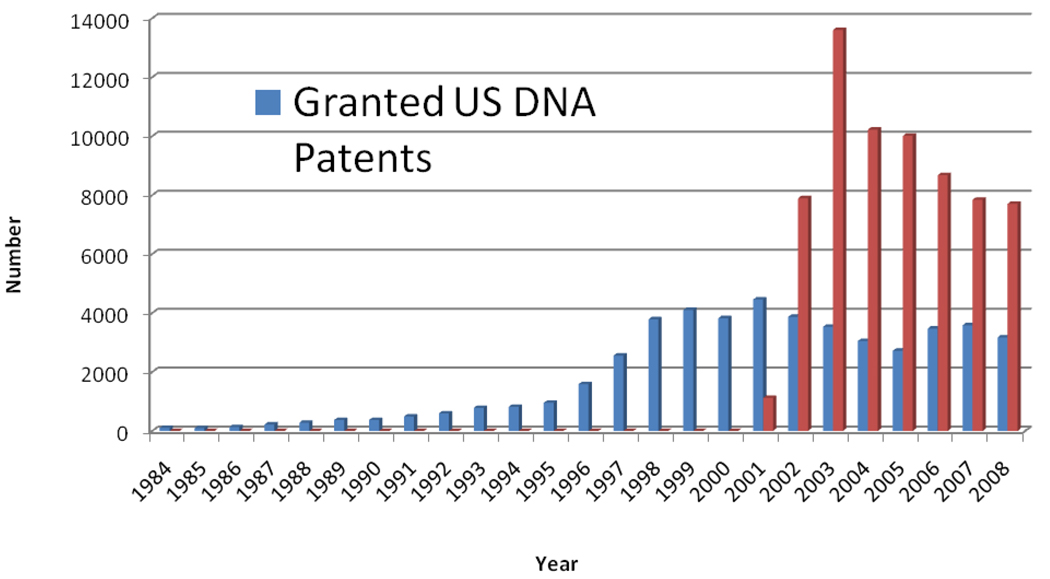
In April 2009, the U.S. Patent and Trademark Office (USPTO) granted the 50,000th U.S. patent that entered the DNA Patent Database at Georgetown University. That database includes patents that make claims mentioning terms specific to nucleic acids (e.g., DNA, RNA, nucleotide, plasmid, etc.) (64). The specificity of many terms unique to nucleic acid structures makes it possible to monitor patents that correspond to and arise largely from research in genetics and genomics. Patents have been a part of the story of the rise of genetics and genomics since the 1970s, and not just because they can be counted but also because science and commerce have been deeply intertwined, one chapter in the story of modern biotechnology in medicine, agriculture, energy, environment, and other economic sectors. The first DNA patents were granted in the 1970s, but numbers surged in the mid-1990s as molecular genetic techniques began to produce patentable inventions. [See Fig 1.]
Figure 1.
U.S. Patents: DNA Patents and Patent Applications by Year, 1984--2008. The DNA Patent Database contains patents obtained by searching the Delphion Patent Database (http://www.delphion.com) with an algorithm posted on the DNA Patent Database website that searches for granted U.S. patents (since 1971) and published applications (since 2001) in U.S. patent classes related to genetics and genomics as well as claims that include words specific to nucleic acids, genetics, and genomics. The year 1984 is the first for which more than 100 granted patents are in the DNA Patent Database. Data from Reference 64.
What Is a Patent? Who Grants a Patent?
A patent is a document issued by a government entity that confers the right to exclude others from making, using, selling, importing, or offering to sell an invention claimed in the patent. That right is enforced by national courts. A patent is, in effect, a license to sue someone for making, using, or selling an invention without permission.
Patent offices grant patents in response to patent applications. The procedural rules differ somewhat, but the criteria for granting patents are broadly similar worldwide. An invention must be patentable subject matter. The U.S. definition is “any new and useful process, machine, manufacture, or composition of matter, or any new and useful improvement thereof” (227). An invention must also meet three criteria for patentability: (a) novelty, (b) nonobviousness (the European term is inventive step), and (c) utility (or in Europe and most other jurisdictions, industrial application). Moreover, a patent must describe an invention in sufficient detail that a “person having ordinary skill in the art” will be able to make and use it without “undue experimentation.” The patent must be “enabling” and the “written description” sufficient.
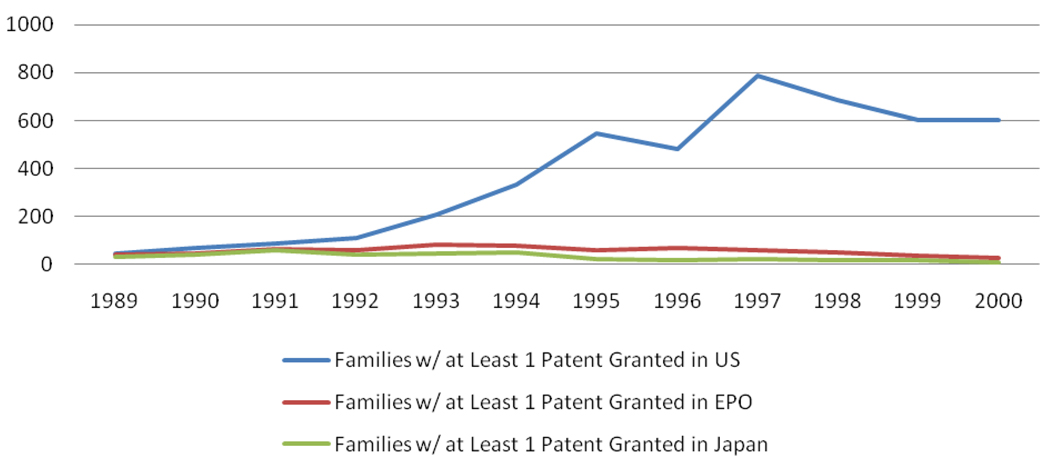
The process for ensuring that these criteria are met is patent examination. International consensus about general patent criteria does not, however, necessarily lead to consistent interpretation and implementation across jurisdictions. Genomics is one of the fields in which interpretation of patent criteria is most divergent, leading to disparate outcomes. The United States grants far more DNA-sequence-based patents (111, 112), for example [See Fig 2], and allows generally broader claims than the other patent offices serving large biotechnology markets in Japan and Europe. These patents are also issued significantly faster in the United States compared to Europe, which is in turn a bit faster than Japan (112, Fig. 7, p. 21).
Figure 2.
Families of Granted Patents in the United States, Europe, and Japan. The figure shows differences among patent offices in families of DNA-sequence-based patents. A patent family is the collection of patent applications and the granted patents arising from a single invention, usually stemming from the same original application. Hopkins et al. (111) “used Thomson Scientific’s GENESEQ and World Patent Index databases to identify patent families claiming human DNA and/or other nucleic acid sequences that were published from 1980--2003. Other data (e.g., legal status of granted patents) were obtained from the U.S. Patent and Trademark Office and the European Patent Office online databases” (111, p. 185). Data from Reference (112) used with permission of the authors.
Differences in patent practice can be important to scientists working in genetics and genomics. In the United States, a patent goes to the first inventor. If patents or patent applications overlap and the first person to invent is in dispute, then the patent office initiates what’s called an interference proceeding, with intricate rules about deciding priority of invention. Interferences are more than twice as common in biotechnology patents than in any other patent class, six times higher than patents on average (140). The United States also allows a year’s grace period from publication of information pertinent to a patent claim, whereas any public disclosure becomes “prior art” that can defeat patent claims in other jurisdictions.
Several international treaties harmonize procedural rules throughout the world. The Patent Cooperation Treaty of 1970 established international practices so that a patent application filed in one jurisdiction can be pursued in others. The 1973 European Patent Convention created the European Patent Office (EPO). The Convention includes some countries that are not in the European Union (e.g., Switzerland, Turkey, and Norway). The EPO can issue a patent valid in signatory countries, but those patents must also be formally recognized by member nations during the national phase, and litigation is, at least for now, entirely in national courts. In December 2009, Ministers of the European Union supported moving toward litigating patent disputes in a trans-European court system, but the idea awaited endorsement from the European Court of Justice and would require a transition to a true European patent (199). In 1976, Africa developed the African Regional Intellectual Property Organization.
The 1995 Trade-Related Aspects of Intellectual Property Rights (TRIPS) agreement committed signatory countries to adopt patent standards mainly modeled on the developed-country model of strong patent protection (191, 192), including patenting of medical products that in many countries had been excluded before and protected only by process patents or not at all (138, 191, 192). Countries agreed to a time line to make their national law TRIPS-compliant, with developed economies first, middle-income next, and low-resource countries given the longest to comply.
Brody reviewed the extensive decade-long debate about biotechnology patenting in Europe that led to the 1998 Biotechnology Directive (34). Gold and Gallochat explained how the Directive became an important element of European patent law that binds national governments to comply with it, despite some resistance and squabbles over some elements (95). The Biotechnology Directive explicitly permits patenting of genes but with somewhat higher thresholds for patentability than U.S. law, for example, requiring that claims on DNA sequences encoding a protein also specify that protein’s function and an industrial application (76, 95).
The upshot for scientists is that patent procedures and rules are generally similar around the world, but there are important differences, and decisions about whether, when, and where to patent often require specialist knowledge; scientists not prepared to do extensive study are well advised to consult with their institution’s technology licensing office or another source of that knowledge.
Two other features of European law are particularly relevant to patents in genetics and genomics. Moral objections are explicitly recognized as a reason not to grant patent rights under European law. EPO also has an administrative mechanism for challenging a patent after it has issued, called opposition, which does not exist in U.S. law. The U.S. patent system has two procedures for re-examination within the patent office, but the grounds for challenge are narrower. These procedural matters can change outcomes. The European opposition process significantly narrowed patent claims pertaining to genetic testing for breast and ovarian cancer, for example, compared to analogous patents in the United States.
Patent reform legislation pending in the United States could reduce some of the differences between rules in the United States and other jurisdictions. Both the House and Senate of the 111th Congress are considering bills similar to one passed by the House of Representatives (but not the Senate) in the 110th Congress (2007--2008). Two provisions particularly relevant to genetic and genomic inventions are (a) shifting from the current “first to invent” U.S. standard to “first inventor to file,” as in the rest of the world; and (b) establishing a mechanism to challenge patent claims closer to the European opposition process.
Why Do Governments Grant Patents?
The constitutional rights granted in the patent clause are not human rights but instrumental rights, or privileges. The constitutional patent clause is an authorization for Congress to give inventors the right to exclude others temporarily in return for making their discoveries public. DNA patents are relatively new, but new technologies are not. DNA patents have been granted under legal rules that accommodated many new technologies of the Industrial Age.
Bugbee traces the first invention patent to Florence in 1421, and the first patent law to Venice in 1474, establishing a process to grant exclusive rights for 10 years in the territory controlled by Venice in return for public disclosure (35B, 155). The English Parliament passed the Statute of Monopolies in 1624, not to create a new right but rather to rein in the king’s power to grant monopolies as sources of income and political patronage (155). Parliament preserved patents of invention but moved adjudication to common law courts (143). Invention patents were intended to promote the collective good, replacing monarchic whim with a principled rule of law.
European patent law informed the debate in a new republic, giving rise to the patent and copyright clause in the U.S. Constitution, which explicitly empowered Congress to “promote the progress of science and useful arts, by securing for limited times to authors and inventors the exclusive right to their respective writings and discoveries” (203). Congress passed a patent statute based on this authorization, and Thomas Jefferson was the first Commissioner of Patents. The last major reform of the patent act took place in 1952. One structural change---the formation of a Court of Appeals for the Federal Circuit (CAFC) to hear patent, international trade, veterans’ benefits, federal contracting, and certain other specialized kinds of appeals---took place in 1982, centralizing appeals to a single court unlike most other civil law appeals that go through regional appeals courts.
Shifting U.S. Jurisprudence
The patent law and structure of the courts have been fairly stable during the “genome era” from 1980 to present, but the interpretation of patent law as it pertains to genetics and genomics changes in response to technology and the real world experience brought to the courts in the form of actual cases. Jurisprudence, like science, is organic and changing, although through a completely different process and on a different time scale.
Patent eligibility
A few cases either pending or recently decided are particularly relevant to genetics and genomics. The CAFC decided a case in September 2009, Prometheus v. Mayo (177), that is being appealed to the U.S. Supreme Court, which has not yet decided whether to accept it. It is not a gene patent case but one about medical testing, that is, administering a drug and measuring drug metabolites to guide treatment. Depending on what is decided and the grounds for the decision, it could have implications for DNA patents, especially for diagnostic uses.
The central feature of the case is what is eligible to be patented (i.e., whether the claimed patent matter complies with 35 U.S.C. 101 (227)). The patent claims were judged invalid by federal district court and CAFC reversed its ruling. The Supreme Court previously took up another medical testing case, Labcorp v. Metabolite, that raised similar issues, but in June 2006 decided its consideration of the appeal had been “improvidently granted” (130). Three justices, led by Justice Breyer, dissented. The district court in Prometheus found Breyer’s dissent “persuasive” when invalidating the patent claims. In its reversal, the CAFC pointedly said Breyer’s dissent is “not controlling law” (177, p. 1082). This tug of war among the three levels of federal courts over the boundaries of what is patentable in a case about medical testing could be decided by the Supreme Court.
The U.S. Supreme Court has heard oral arguments and will soon decide another landmark case about “business methods” patents that is, again, about what is eligible to be patented. Bilski v. Kappos addresses patenting methods used by financial hedge funds (30), not genomics, but it is being closely watched because it could shift the line or set new rules for deciding what methods can be patented, such as those that correlate DNA sequences with traits, genetic risks, or diseases.
Obviousness
The Board of Patent Appeals and Interferences decided Ex parte Kubin in 2007 (77), a case turning on technologies for cloning genes from known protein amino acid sequences. Until that decision, a nucleic acid sequence was deemed “nonobvious” for patent purposes until and unless its sequence had been specified, under a CAFC rule from a 1995 case (118). Yamanaka asked if Kubin were “a nail in the coffin for DNA sequence patents?” (225), but the case actually only renders vulnerable those patents based on having cloned a gene for a protein whose structure was already known as of September 2000. It does not affect DNA sequence patents for newly found genes whose functions were not previously known, the majority of inventions in the genomic era. Kubin was based on a new Supreme Court precedent that gave more discretion to the courts in deciding “obviousness” (129).
Court discretion in injunctive relief
Goldstein notes that another U.S. Supreme Court decision, eBay v. MercExchange (67), could affect genetic diagnostics, particularly tests that involve multiple components (96). The issue was whether a patent holder could block an alleged infringer from making and selling a complex invention, only a small part of which was covered by the patent. The district court said no, the infringement did not justify an injunction; the CAFC said yes, both district and appeals courts more or less automatically denying and then granting an injunction, respectively. The Supreme Court said that courts should exercise discretion in granting injunctions and set a four-way test. Before this case, those enforcing patents could rely on getting an injunction, and this case weakens the incentive for those holding patents on small components to hold out or litigate. In genetics and genomics, this applies to technologies such as microarrays that use many DNA sequences and to complex instruments that embody many different patented components.
Who Owns DNA Patents?
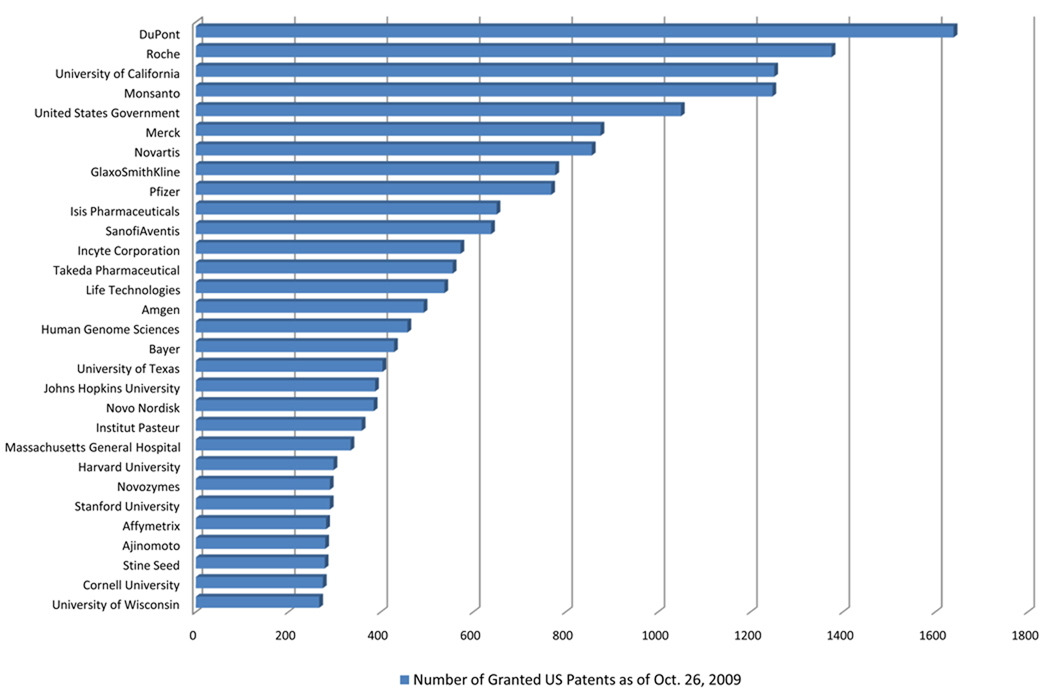
Figure 3 shows the top 30 institutions holding patents in the DNA Patent Database. Among them are
Agribusiness and chemical companies (Monsanto and DuPont)
U.S. Government (largely attributable to the large intramural research program at the National Institutes of Health)
Public and private universities (Universities of California and Texas, Johns Hopkins, Harvard, Stanford, MIT, etc.)
Pharmaceutical firms (Novartis, Glaxo SmithKline, Pfizer, Merck, SanofiAventis, Takeda, Bayer, Novo Nordisk, Lilly, etc.)
Established biotechnology firms (Genentech, Amgen, Genzyme, ISIS, etc.)
Firms created to exploit genomic technologies (Incyte, Human Genome Sciences, etc.)
Instrumentation and DNA chip firms (LifeTechnologies, Affymetrix, Becton, Dickinson, etc.)
Academic research institutes (Institut Pasteur, Salk, Scripps, and Ludwig Institutes, Cold Spring Harbor Laboratories, etc.)
Hospitals with research units (e.g., Massachusetts General Hospital)
Figure 3.
Top U.S. DNA patent holders. The authors compiled a list of assignees with at least 100 patents, combined different names for the same assignee, and updated names to reflect corporate mergers and acquisitions. Patent counts are from the Delphion Patent Database for U.S. patents granted as of October 26, 2009, using the DNA Patent Database algorithm (64). Data from Reference 64.
The mix of large and small, new and old firms would be found in other kinds of patents. The number of nonprofit institutions (government, universities, research institutes, and hospitals), however, is highly unusual. Overall, fewer than 3% of U.S. patents are owned by academic institutions, compared with 39% in a comprehensive analysis of all DNA patents granted in the United States from 1980–1993 (S. McCormack, R. Cook-Deegan, unpublished data). The subset of DNA patents claiming sequences corroborates this pattern, with public sector (nonprofit) owners accounting for roughly half through the mid-1990s and for more than a third from 2000–2003 (112, Fig. 8, p. 23). Academic institutions are a far more important patent-holding constituency in genetics and genomics than in general.
DNA PATENTS INTERACT WITH SOCIAL AND POLITICAL CONCERNS
Human genetics and genomics differ from many other fields of research and development (R&D) in the nature of the downstream products and in a strong general interest in and concern about how the science is done, how it is applied, and how fairly its benefits are distributed. The research itself touches human lives directly, and human beings or their cells are often the objects of research. Ownership of data, materials, and control spill over into Who owns this? questions that are more pointed with reference to genes than for computers or cell phones. Fairness and access are important values in health and health care.
Just weeks after the U.S. Supreme Court decided Diamond v. Chakrabarty in 1980, thereby permitting patents on living organisms, the General Secretaries of the three largest U.S. religious denominations jointly signed a letter to President Jimmy Carter raising questions and concerns:
Who shall determine how human good is best served when new life forms are being engineered? Who shall control genetic experimentation and its results which could have untold implications for human survival? Who will benefit and who will bear any adverse consequences, directly or indirectly?
These are not ordinary questions. These are moral, ethical, and religious questions. They deal with the fundamental nature of human life and the dignity and worth of the individual human being.
With the Supreme Court decision allowing patents on new forms of life---a purpose that could not have been imagined when patent laws were written---it is obvious that these laws must be reexamined. (Randall C., National Council of Churches, Mandelbaum B., Synagogue Council of America, Kelly T., U.S. Catholic Conference. Letter to President Jimmy Carter. See Reference 181)
Media Attention and Policy Reports
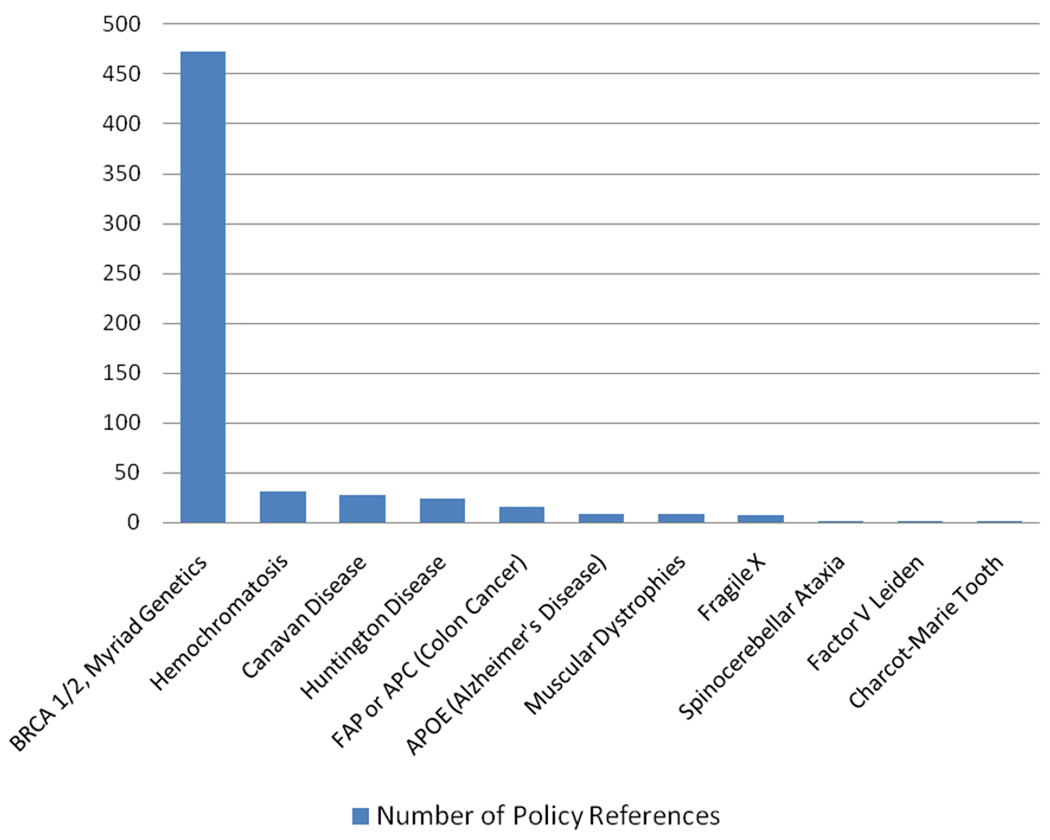
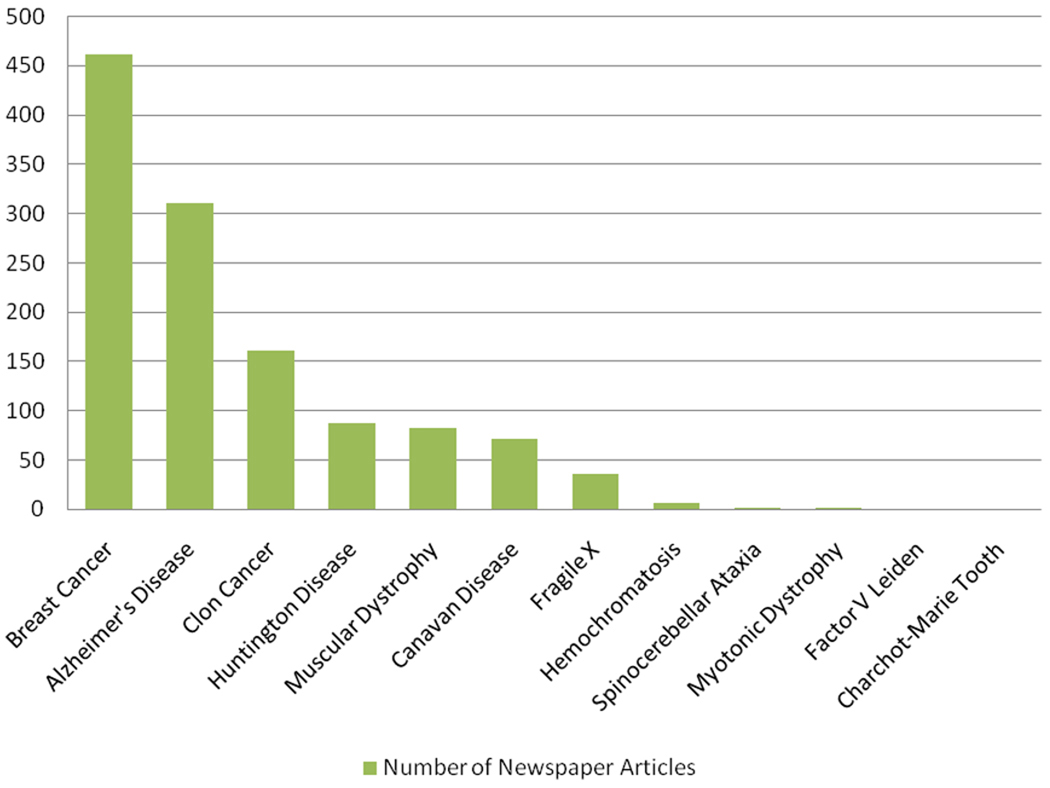
Patents in genetics and genomics have stirred controversy. A media content analysis of gene patent controversies in English language newspapers showed that patents on the BRCA1 and BRCA2 genes associated with inherited risk of breast and ovarian cancer stood out as by far the most salient (Figure 4a). Media coverage was predominantly negative even in the United States, where the patent holder, Myriad Genetics, was located. In Australia, the United Kingdom, and especially Canada, where Myriad threatened patent enforcement, coverage was overwhelmingly negative (38).
Figure 4.
Newspaper stories and policy report references to genetic conditions, genes, and related controversies. References to gene patents in English language newspapers and policy reports, according to gene or to company. Caulfield and colleagues (38) searched English language media in Australia, Canada, the United Kingdom, and the United States for the period 1994--2006 for stories about gene patents. (a) The number of newspaper articles that mentioned specific conditions, genes, or controversies was counted. (b) In another article, Caulfield and colleagues (39) searched for explicit references to specific gene patents and firms in English language policy reports that addressed gene patenting from 2002--2006. Shown are the number of times specific patents and firms were mentioned in those reports (excluding irrelevant or synonymous uses of terms). The number of references in policy reports to Myriad Genetics and BRCA1/2 are combined here but were reported separately in the two Caulfield et al. publications (38, 39); references in policy reports to various muscular dystrophies were also reported separately but combined here. Data from References 38 and 39 used with permission.
Caulfield et al. (39) reviewed mentions of patent controversies in 18 policy reports from around the world and again found that the BRCA patent controversy vastly outstripped others (Figure 4b). The Ontario government’s 2002 report (166), for example, was clearly fueled by the highly public Myriad Genetics controversy, and laws were passed in France and Belgium under the shadow of BRCA. Gold & Carbone’s (94) case study shows how tensions over gene patenting moved from smoldering concern to burst into controversy and led to deliberate disregard of Myriad’s patents as a matter of policy and political strategy. Shobita Parthasarathy paints a similar picture of push-back in the United Kingdom, where the National Health Service was Myriad’s main potential customer (173). A lawsuit brought against Myriad and codefendants in May 2009 was brought not by a competitor company but by a consortium of medical organizations and individual plaintiffs and was sponsored by the American Civil Liberties Union (20).
BRCA might be the biggest bone of contention, but policy attention to DNA patents predated the introduction of BRCA genetic testing by Myriad Genetics. Indeed, it went back to the early 1980s, with the emergence of biotechnology and the 1980 Chakrabarty decision. The congressional Office of Technology Assessment (OTA) published Impacts of Applied Genetics in 1981, Commercial Biotechnology: An International Analysis in 1984, Patenting Life in 1989, and Biotechnology in a Global Economy in 1991 (160--163). Each OTA report had at least a chapter on patents, and the 1989 report was entirely devoted to the subject. Another OTA report directly focused on DNA patents, The Human Genome Project and Patenting DNA Sequences. It was approved for final revision and publication in 1994 (164), but Congress defunded OTA in 1995 and that report was never published (135).
Policy reports about DNA patents were produced in many other countries. The United Kingdom’s Nuffield Council on Bioethics and the Organization for Economic Cooperation and Development in Paris issued reports in 2002, the United Kingdom Public Health Genetics Unit in 2003, both the Danish Council of Ethics and the Australian Law Reform Commission issued reports in 2004, and the World Health Organization in 2005 (22, 58, 159, 167, 202, 224).
In 2006, the National Research Council (NRC, the operational arm of the U.S. National Academy of Sciences) issued a report on patenting in genomics and proteomics (157). The Secretary’s Advisory Committee on Genetics, Health, and Society (U.S. Department of Health and Human Services) then initiated a task force on the impact of patenting and licensing on clinical access to genetic testing. That initiative was intended to complement the 2006 NRC report, which had touched on diagnostics but mainly emphasized impacts on research.
New Laws and Legislative Activity
Members of Congress and foreign parliaments have made statutory changes influenced by concerns about DNA patents. France and Belgium created compulsory licensing authorities that were influenced directly by concerns about breast cancer genetic testing (207, 208, 209, 214), and Switzerland has a somewhat different mechanism for compulsory licensing that could be applied to genetic diagnostics and therapeutics (89).
In the United States, several bills on DNA patents have been introduced since 1992, although none has become law. In March 1992, Senator Mark O. Hatfield proposed a 3-year moratorium on patents claiming patent rights to any “human tissue, fluid, cell, gene or gene sequence (genetically engineered or otherwise)” until Congress could consider a series of reports (206). Representatives Lynn Rivers and David Weldon introduced a bill, HR 3967, in March 2002 to exempt research and genetic diagnostic use from patent infringement liability (meaning such uses would be permitted) and to mandate early disclosure of DNA sequence information in patent applications (204). Reps. Xavier Becerra and David Weldon introduced HR 977 in February 2007, a short bill stipulating that “no patent may be obtained for a nucleotide sequence, or its functions or correlations, or the naturally occurring products it specifies” (205). The introduction of the Becerra--Weldon bill followed the efforts of novelist Michael Crichton and legal scholar Lori Andrews, who teamed up following publication of Crichton’s penultimate novel Next in 2006. Next centered on corporate corruption involving gene patents, alluded to Myriad Genetics and breast cancer genetic testing, and included a nonfiction appendix that called for an end to gene patents and repeal of the Bayh--Dole Act (56). Crichton’s New York Times February 2007 OpEd, “Patenting Life,” began, “You, or someone you love, may die because of a gene patent that never should have been granted in the first place” (57). This was strong stuff apt to get the attention of those making policy decisions.
The Secretary’s Advisory Committee on Genetics, Health, and Society (SACGHS) approved a set of recommendations about patenting, licensing, and use of DNA patents relevant to clinical genetic testing in October 2009, and a full report was approved in spring 2010 (190, 190A).
THE EMERGENCE OF GENOMIC PATENTS
Genetics and Genomics: Born into Biotechnology
From the beginning of the 1980s, hot science in molecular biology and the promise of wealth and jobs from biotechnology grew hand-in-hand. Molecular genetics, biotechnology, and expectations of economic development were parts of a package. Human genetics and genomics grew into prominence as part of this history. Human genetics and genomics fit squarely into Pasteur’s Quadrant, where research was both conceptually and scientifically important, but at the same time had obvious and foreseeable practical benefit (198), and often commercial value.
June 1980: Diamond v. Chakrabarty
In June 1980, the U.S. Supreme Court handed down a 5--4 decision in Diamond v. Chakrabarty (60, 124). The U.S. patent office had denied General Electric’s patent application, because it claimed a life form, a bacterium selectively bred to metabolize petrochemicals and designed to digest oil spills. The Supreme Court determined that this was an invention eligible for patent protection. This case was decided amid a very public national discussion about recombinant DNA, and fears of biohazards arising from splicing genes into living organisms (86, 127), but also as successes in gene cloning were showing the promise of biotechnology.
While Chakrabarty’s modified Pseudomonas bacterium was not made from recombinant DNA, the Supreme Court decision was nonetheless taken as a strong signal that products and organisms made that way were patentable. Universities, pharmaceutical firms, and start-up companies with a stake in the nascent biotechnology “industry” weighed in with briefs, generally favoring extending patent rights to cover Chakrabarty’s bacterium (124). Patents on underlying methods, such as Cohen--Boyer cloning and Axel cotransformation, had been under examination for several years, and the first gene patent applications, including insulin and growth hormone, had already been filed.
October 14, 1980, was significant in the interwoven histories of recombinant DNA, DNA sequencing, and commercial biotechnology. Walter Gilbert and Frederick Sanger won the Nobel Prize for their respective DNA sequencing methods, and Paul Berg for his work on recombinant DNA. Herb Boyer, whose UCSF laboratory was deeply involved in DNA cloning technology, was also a cofounder of Genentech with venture capitalist Robert Swanson. Boyer was huddled with others for Genentech’s initial public offering (first sale of publicly traded stock) and recalled that day’s San Francisco Chronicle: “…the headline was ‘Genentech Jolts Wall Street’ and underneath is a photo of Paul Berg, ‘Berg Wins Nobel Prize’” (93). The modern era of molecular genetics juxtaposed Nobel-quality science with a big business story.
DNA Patents before Chakrabarty
U.S. patent 3,615,654 was arguably the first nucleic acid patent, covering a method for treating cells with liquid ammonia and thereby changing protein and nucleic acid composition of the cells; RNA was explicitly mentioned in the final three claims (25). Similarly, most early DNA patents were about foodstuffs or chemical treatment of cells. Very few of the 159 patents granted during 1971--1980 that mention nucleic acids actually used molecular biological methods. A few, however, presaged the tide of DNA patents soon to come from molecular biology. Peter Gilham and Herbert Weith of Purdue University, for example, secured a 1973 patent that was in effect a method for DNA sequencing, although it never proved practical (92, 189). Johns Hopkins University got a 1977 patent on nucleic acids that induced interferon production (201). Yet such patents did not provoke a public debate about patents and biotechnology. That changed in December 1980.
New molecular genetic technologies took center stage with US Patent No. 4,237,224, the first of three patents issued to Stanley Cohen of Stanford University and Herbert Boyer of the University of California, San Francisco, covering recombinant DNA methods (45). Just 10 days later, on December 12, 1980, the U.S. Congress passed the Bayh--Dole Act. These events were largely independent but carried along by the same policy stream.
Cohen--Boyer Patents and Shifting Norms of University Patenting
The Cohen--Boyer patent culminated a seven-year story (114, 185, 186). Stanley Cohen of Stanford and Herbert Boyer of UCSF met in a Waikiki Beach café in November 1972 to brainstorm about constructing plasmids, or circular DNA molecules that replicated inside bacteria, from pieces of DNA derived from different organisms. They published the first such chimeric plasmid in November 1973, with Annie Chang and Robert Helling (46).
Niels Reimers at Stanford saw an opportunity to patent and license a powerful new technology with obvious commercial implications. Reimers was trying to develop Stanford’s patenting and licensing portfolio into a spur for innovation and a source of university income. The decision to patent was fateful in four respects. It broke ground in patenting a method central to molecular biology and its applications, it raised the question of who would control patent rights from federally funded inventions, it led to a novel licensing strategy, and that licensing brought in a quarter billion dollars in revenue. Among molecular biologists, it also signaled a norm shift. DNA sequencing methods developed around the same time by Maxam and Gilbert at Harvard, and by Sanger and Coulson in Cambridge, UK, were not patented but certainly could have been. The 1973 Purdue patent on a DNA method showed such a patent could be obtained. Yet Walter Gilbert and Fredrick Sanger, interviewed years later, said they did not consider patenting their DNA sequencing methods, because they conceived of them as basic research methods (91; R. Cook-Deegan, personal communication). Likewise, the pBR322 plasmid, which became a workhorse for gene cloning for many years, was an immensely clever piece of engineering by UCSF’s Fernando Bolivar and Ray Rodriguez, but it was not patented. Yet the method of recombinant DNA that it embodied was patented at the same university during the same period. Norms among scientists and universities about what to patent were shifting, but they were not uniform or consistent.
Start-ups, products, and revenues helped push toward commercial applications of molecular biology. Stanford eventually generated 468 licenses covering 2,442 products before its Cohen--Boyer patents expired; the $255 million in revenues generated for Stanford and the University of California before the patents expired in 1997 tapped the $35 billion in sales of recombinant DNA products (78, esp. pp. 1803--5). Stanford did not seek licenses from academic research institutions, creating a de facto exemption for academic research; its licenses were based on the production of commercial end products. Stanford kept royalty rates and up-front payments relatively low to encourage licensing and discourage commercial licensees from litigation. Columbia University used a similar strategy a few years later with its cotransformation recombinant DNA technology, which generated an estimated $790 million in revenues (49).
The Bayh--Dole Framework
The National Institutes of Health (NIH) and the National Science Foundation (NSF) funded the research that produced the Cohen--Boyer patents. This raised the question, Who would own the patent rights: the inventors, their universities, or the federal government that funded the research? As it pursued patent applications, Stanford sought permission from the NIH and NSF to retain the patent rights. The NIH director, Donald Fredrickson, sent a letter to many university presidents and administrators seeking counsel (85). Both the NIH and NSF had been moving toward giving patent rights on federally funded inventions to their grantees and contractors, and not surprisingly, that policy was espoused in most of the replies to Dr. Fredrickson’s letter (156). The NIH gave Stanford permission to patent but stipulated that the technology should be licensed nonexclusively and broadly so that it could be widely adopted, with exclusive licensing only if that failed or could otherwise be justified (84). The NIH took a similar tack with Columbia University for its cotransformation patents (141).
On December 12, 1980, Congress passed Public Law 99--517, which conferred on grantees and contractors the option to seek patents (228, as implemented in Ref. 229). This statute gave institutions, such as universities, nonprofit research institutes, and small businesses, the first option to acquire patent rights on inventions arising from federal funding. This was later extended by executive order to larger firms (182). The statute became known as the Bayh--Dole Act, for its Senate sponsors, Birch Bayh and Robert Dole. The purpose of the Act was to “use the patent system to promote the utilization of inventions arising from federally supported research” (228, Sec. 200). It set presumptive ownership rules, giving grantee or contractor institutions the right to retain title to inventions arising from federal funding, and it created much more consistent policies among the many federal funding agencies.
The original Cohen--Boyer patent application was filed in 1974, and the first patent was granted on December 2, 1980. During this period, federal agency practices about patent ownership were inconsistent. While the Cohen--Boyer patents involved the NIH and NSF, whose policies were largely consonant, other research might entail funding from the Department of Defense or the Department of Energy (DOE), where government ownership of resulting patents was more often the norm. Research institutions had to negotiate patent rights with each funding agency while they were applying to the patent office for the patents, increasing the cost and complexity. By making the process simpler and more consistent, the Bayh--Dole Act encouraged research institutions to patent and license their inventions.
The idea behind the Bayh--Dole Act took root in 1978. It was passed in a lame duck session of Congress, the month after Birch Bayh had lost his re-election campaign, and partly as a favor to the departing Senator (197). One major argument for the new law was that without the incentive to research institutions, the federal government was leaving inventions to languish. Bayh--Dole advocates cited the 28,000 patents owned but rarely licensed by the U.S. Government. The evidence behind this claim was flimsy since most of the patents came from defense research in which contractors had declined exclusive rights (70). The stronger arguments were about consistency and simplicity in the rules for patenting inventions from federally funded research; and of course research institutions would surely like the money. The crisis of confidence about U.S. economic competitiveness also became a rallying cry for the bill’s proponents, with the Bayh--Dole incentives being a way to tap the innovative value of America’s great research universities (29).
The Bayh--Dole Act was less the prime cause of a revolution and more the codification of emerging practices. While Cohen--Boyer recombinant DNA, Axel cotransformation, and other key DNA technologies arising in academic research predated Bayh--Dole’s enactment, ownership of the relevant patents was similar to what would have happened after it passed. The Bayh--Dole Act simplified the rules just as molecular biology was proving valuable in biotechnology and set the stage for academic--industrial mutualism in genetics and genomics. The lucrative licensing of recombinant DNA technologies at Stanford, the UC system, and Columbia became an object lesson for other universities.
These technologies were never in any danger of languishing in academic or government laboratories (50, 146). They did represent successful technology transfer from university research to commercial application, and one part of the “transfer” attracted considerable attention: the flow of dollars to universities based on their patent licenses. This was not the stated rationale of the Bayh--Dole Act, but the policy did reward socially useful activity at the responsible institutions and also compensated the inventors. The just deserts rationale was not prominent in the Bayh--Dole debate, but it could have been and should have been an explicit basis for policy choice, based on evidence more credible than the “languishing invention” arguments (49).
In the 1980s and 1990s, many more universities developed technology licensing offices, and the number of patents to academic institutions grew dramatically, particularly in the life sciences (44, 145, 146). Having staff and expertise increased the propensity to patent since the infrastructure was in place. Scientists saw benefit in translating their discoveries into real-world applications, and some of the resulting money came back to them and to their institutions, supporting research and education. Jobs and wealth grew out of such translational activities.
Increasing links between universities and industry provoked a debate, often framed as a fight for the soul of academic research, a dichotomous choice between revenues, commercialization, and economic growth, on one hand, and disinterested pursuit of pure science as a public good, on the other. Many faculty and administrators at universities and many scientists and corporate officers in industry rejected this dichotomous frame, however, and worked to make both hands clap together. Policy makers wanted the jobs and wealth from biotechnology, but they also wanted neutral and objective health research funded by taxpayers.
Eric Campbell, David Blumenthal, and their colleagues surveyed scientists in 1985, 1995, and 2007. They concluded that “relationships [with industry] are most common among productive, senior faculty members who contribute substantially to their research community,” as measured by publications, engagement with national organizations, and other indicators (226, p. 1822). More than half those in the 2007 survey had some industrial association. Industrial partners deliberately choose to work with conspicuously productive researchers of international stature. In the successive surveys, reports of trade secrets increased, both over time and with the degree of industry funding, as did reports of publication delay. But counter-intuitively, industry funding for research dropped as a fraction of total funding among those surveyed. Among the subset of scientists in biotechnology-related fields, “in 1985, 23% of faculty members … reported that they were principal investigators on research projects funded by industry, as compared with 21% in 1995 and 17% in 2007,” with a parallel drop in funding from 7.4% of their total research budgets in 1985 to 6.1% in 2007 (226, p. 1821). Given that the use of molecular biology in industry grew continually, why the drop in academic funding in the survey? Firms internalized R&D; small start-up firms conducted some R&D previously done at academic centers; clinical research moved to private-contract research organizations; and universities outside the United States grew as an alternative.
The classic technology transfer stories entail a research discovery that is transformed into a product or service through a broad and complex R&D network that includes private firms. Academic institutions generally do not make drugs or scientific instruments beyond the prototype stage. In the process of making academic research results useful to commercial partners, however, patenting and licensing are generally less important than publications and “open science” (47). In particular cases, however, patents are important. Patent rights are a mechanism for handing off a discovery with rights that enable subsequent private R&D investment, but they also impose requirements to keep information private, at least until patent applications are filed. Academic scientists can be links in a private R&D chain where Mertonian norms of open science collide with demands to keep data proprietary. In surveys of those involved in proteomics and genomics, Walsh and colleagues found patents well down on the list of impediments to innovation, and few scientists checked whether they might be infringing patents in their research (219, 220). The degree of friction caused by patents per se was less than that associated with exchanging research materials.
As the new institutional framework became established, several books and articles raised the specter of corruption as universities intensified their ties with private industry (31, 97, 128, 176, 221). Taking a more pragmatic tack, economic theorist Richard Nelson made note of the unique value of open science as practiced at academic institutions (158), making a plea for the social mission of the university and joining legal scholar Rebecca Eisenberg in calling for “reasserting the value of public science as broadly valuable … not limited simply to the products or technologies it spawns” and that can be patented and licensed (73, p. 1392).
Patenting Genes
Several technologies were particularly conspicuous among the early DNA patents, and in biotechnology more generally, among them: recombinant DNA cloning, DNA and RNA sequencing, synthesis of DNA and RNA molecules, polymerase chain reaction (PCR), cell fusion techniques for making monoclonal antibodies, and computational tools to analyze data from molecular genetic analysis.
In addition to the lucrative recombinant DNA method patents noted above, some of the most valuable early DNA patents claimed DNA molecules that specified the amino acid sequence of proteins with known therapeutic value (e.g., insulin or growth hormone) or were patents covering methods or sequence variants (mutations) associated with diseases. These are often referred to as gene patents, although that term is used in many different ways. Other DNA patents covered basic methods used in research, production of medicines, vaccines, scientific instruments for studying DNA, and algorithmic methods for interpreting genomic data. Indeed, the majority of patents in the DNA Patent Database do not make claims about specific DNA sequences, but considerable attention has fallen on DNA sequence patents.
First Generation Gene Patents
First-generation gene patents were valuable because they enabled production of therapeutic proteins such as insulin, growth hormone, tissue plasminogen activator, and blood clotting factors. Those patents covered DNA sequences discovered by cloning a gene for a known protein. Cloned DNA constructs were then inserted into cells to produce the proteins faster, in greater amounts, with higher purity, and at lower cost than previous methods that relied on extraction and purification of protein from massive amounts of pancreatic tissue, collected pituitary glands, or pooled blood collections. Patent protection was valuable because of the long and costly road that followed gene discovery, from scale-up for commercial production, proof of safety and efficacy in clinical studies, to sale and distribution in medical markets. The substantial investments in these costly developmental stages could be recouped through high prices on final therapeutic proteins-as-drugs because of the patent protection.
Having a patented DNA sequence blocked competitors from making the same protein therapeutic by recombinant DNA. Gene patents were in effect an extension of the small-molecule pharmaceutical business model, with strong patent protection of the DNA that encoded a therapeutic protein rather than a patent on the drug molecule itself. Courts throughout the world adopted this same rationale, based on patents covering “isolated” DNA molecules encoding valuable therapeutic proteins. [Indeed, insulin and growth hormone were also regulated as drugs by the Food and Drug Administration (FDA), for historical reasons, although most later products were treated as “biologics” in a different part of the FDA with somewhat different rules (122).] Several early gene patent stories are reviewed in the first part of Baruch Brody’s classic trilogy of articles on biotechnology patenting in the United States and Europe (32--34).
Recombinant DNA technology was crucial in making valuable therapeutic proteins used as drugs. Insulin was the first recombinant DNA product approved for marketing as Eli Lilly’s Humulin®. It was first approved in the United Kingdom and then by the USFDA in 1982 (9). The final product drew on R&D at the University of California, San Francisco (UCSF), biotechnology start-up Genentech, and established pharmaceutical firm Lilly, which already dominated the U.S. market for insulin but foresaw future shortages of insulin extracted from animal pancreas. Stephen Hall’s 1987 book Invisible Frontiers recounts this lively story (99), which Sally Smith Hughes brings up to date in her forthcoming book on the birth of biotechnology in the San Francisco Bay Area (115).
The early history of biotechnology is suffused with patent conflicts. Six lawsuits among UCSF, Genentech, and Lilly erupted over insulin, for example. These were consolidated into a single case tried in federal district court in Indiana (183) and appealed to the Court of Appeals for the Federal Circuit (184). Science observed, “this vicious fight centers on a landmark discovery by UCSF biologists at the dawn of the biotechnology era: the first successful cloning of the rat insulin gene” (136, p. 1028). Thus, the first product of recombinant DNA was the subject of litigation that cost over $30 million and lasted until 1997, two decades after the cloning experiments, with the final appeal decided the same year the Cohen--Boyer patents expired (136, 137). In the end, UCSF’s claims to human insulin were not upheld, and Lilly did not need to pay back royalties. The Court did not invalidate claims to rat insulin, but it did decide that the claims did not extend to human insulin---and there was no big commercial insulin market for diabetic rats. The UC v. Lilly case centered on a technological landmark in gene cloning, and became a legal landmark in its own right: It set precedents in patent-office examination of DNA sequence patents, leading to greater specificity in the “written description” of such inventions and raising the threshold to show “credible, substantial and specific” utility (61--63).
The cloning of somatostatin, insulin, and growth hormone began a string of products derived from applying molecular biology to products and services, initially in medicine and agriculture. These early successes opened a floodgate. The number of DNA patents grew. Among them, gene patents attracted particular attention.
Studies of Gene Patents and Litigation
Kyle Jensen and Fiona Murray linked DNA sequences claimed in patents to RefSeq and GENE databases. They found sequences from 4,382 of the 23,688 known genes in those databases (20%) were mentioned in a patent claim (120). The single largest collection in this dataset belonged to Incyte, most of whose patents were for sequence-based probes. Michael Crichton’s assertion in The New York Times that “one-fifth of the genes in your body are privately owned” overstated the case by a long margin (57). The strategy produced an undercount, missing some “gene patents” that did not make claims on DNA sequence in the way sampled. More important, the claims on sequences in the data set ranged widely from full-length genes encoding valuable proteins, to diagnostics, to claims on probes or research tools that did not confer exclusive ownership of a gene in any meaningful sense.
Jordan Paradise and a team of colleagues from science and law reviewed 1,167 claims from 74 patents on genes associated with nine genetic diseases. Their team assessed whether patent criteria were met (170), essentially an attempt to evaluate the adequacy of the U.S. patent examination process for gene patents of clinical relevance. They concluded that 448 (38%) of claims had a problem. For instance, they found that patents often claimed far more than had actually been invented. While indicating there might be a problem, such a study has no legal authority, however, and the only way to verify the extent of the problem would be either to re-examine these patents or to challenge such patents in infringement litigation.
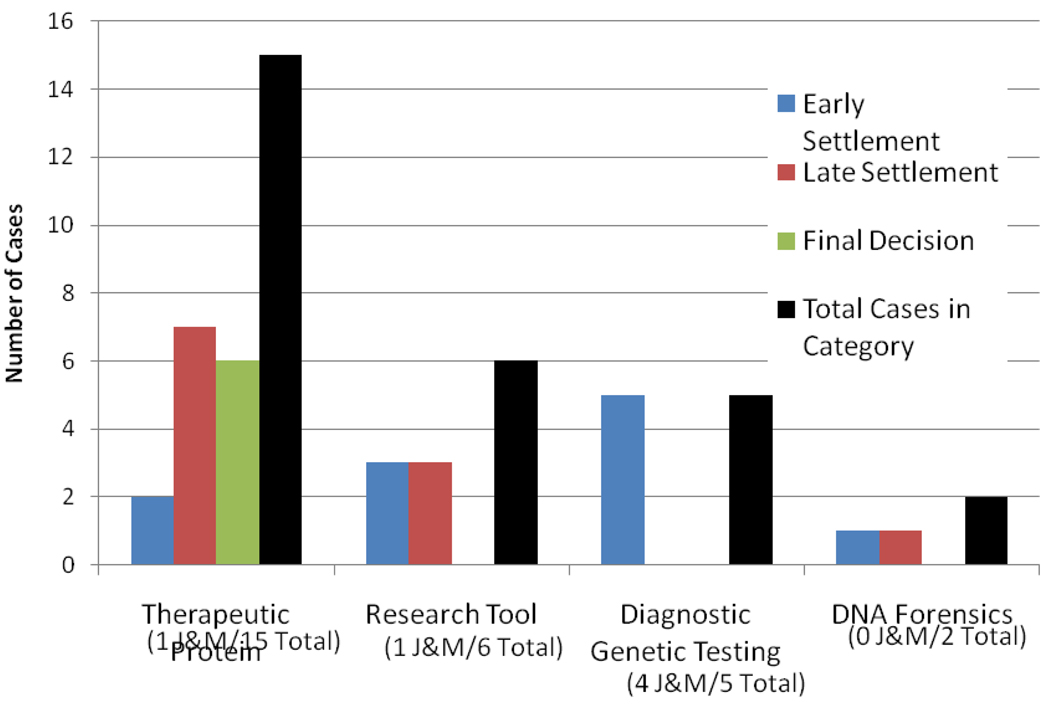
Legal scholar Christopher Holman created a database of gene patent lawsuits in the United States (108, 109). He found 31 cases through early 2007. The frequency of litigation was comparable to other domains of patent litigation. Within DNA patents, many more suits involving sequences encoding therapeutic proteins actually went to trial, whereas all the cases in diagnostics settled very early and none went to court. The March 2010 ruling by Judge Robert Sweet that invalidated patent claims on BRCA genes was the first ruling in a diagnostic gene patent infringment case(21A). Only seven of Holman’s thirty-one cases involved patents identified in the Jensen and Murray data set, and none of those were decided in favor of the patent holder. This study suggested that whereas some therapeutic gene patents had been fully litigated and tested, in court, claims pertinent to diagnosis had not, until the BRCA case. This is due in part to the cost of litigation, which is generally high stakes, with cases often costing at least a million dollars. Perhaps the cost of litigation can be justified when dealing with billion-dollar therapeutic proteins, but the enforcement of patents for diagnostics has generally been mediated by simple letters notifying laboratories that they might be infringing patents (notification letters) or letters to cease and desist from testing. Such letters have generally sufficed to drive university and reference laboratories operating on low margins out of the market rather than challenge patent claims.
THE DIVERSITY OF DNA PATENTS: PATENT STORIES
Studies of aggregate patent data inform debate, but the full diversity of ways in which patents---and their absence---influence technologic innovation also comes through in more detailed narratives of specific cases. The following summaries flesh out the aggregate statistics recounted above.
Erythropoietin
The story of erythropoietin (Epo) includes a multibillion dollar therapeutic protein, the rise of Amgen as a company built on recombinant DNA technology, and three waves of patent litigation. H. Franklin Bunn observed in The New England Journal of Medicine that “recombinant human erythropoietin is arguably the most successful therapeutic application of recombinant DNA technology to date” (36, p. 1901).
Amgen was founded on a business model of sequencing genes encoding protein therapeutics (32). Epo, a hormone that stimulates production of red blood cells, promised to be useful in treating anemia associated with kidney disease, diabetes, and some cancers (100). Amgen cloned the gene for Epo and developed EPOGEN, which generated $663 million in net sales in the third quarter of 2009. A second-generation, modified Epo product, Aranesp®, generated another $685 million for Amgen that quarter. Epogen® and Aranesp® are protected by U.S. patents that expire in 2012--2015 and 2024, respectively. In Europe, the patents expire in 2010 and 2014 (81). Epogen® alone has produced $25 billion in sales since 1989 (174). In non-U.S. markets, excluding Canada, Aranesp® faces competition from biosimilars but retains a 53% market share (178).
Despite Amgen’s recent difficulties with disappointing trial results and allegations of concealing negative trial results (218), the litigation history illustrates the power of patents in biotechnology. Amgen won a make-or-break patent race with Genetics Institute in the late 1980s. In 1987, the Genetics Institute got a patent on purified EPO and a liquid chromatography--based process for producing it. A few months later, Amgen received a patent claiming the genetic sequence for erythropoietin and a recombinant DNA process for manufacturing it (32). Amgen sued the Genetics Institute and its American licensee Chugai Pharmaceutical Corp. Initially, a Massachusetts federal district court found that Amgen and the Genetics Institute each held valid patents that mutually infringed (10). As Nature wrote, “Observers expected that the [Court of Appeals for the Federal Circuit] in Washington, DC, would uphold the Massachusetts ruling and that the continued stalemate would force the companies to cross-license” (90, p. 99). The Court instead upheld Amgen’s patents and invalidated those from Genetics Institute (11). Amgen gained exclusive rights to make Epo in the American market and saw its stock rise 12% in one day. Genetics Institute paid $14 million in damages (32).
The next challenge came from Transkaryotic Therapies (TKT) and Hoechst Marion Roussel (HMR). They produced Epo by inserting promoters adjacent to endogenous EPO genes to work around Amgen’s patents (32). TKT claimed its method did “not require knowledge of the gene sequence” (217, p. 532). In the United States, Amgen prevailed as TKT and HMR were enjoined from infringing Amgen’s patents (12, 81). In the United Kingdom, the House of Lords, which hears patent appeals, reached the opposite decision (126). Amgen ceased marketing Epogen® in Europe (81).
A third wave of patent litigation began in 2009, concerning pegylated Epo, a formulation of Epo that slows protein degradation and extends time between doses. Amgen sued Hoffmann--La Roche in 2005, and Roche launched a countersuit. On September 15, 2009, the U.S. Court of Appeals for the Federal Circuit remanded the case to the District of Massachusetts for retrial (13). As of December 2009, Roche cannot import its infringing product into the United States (17).
Cloning and patenting erythropoietin and modifications of it have produced Amgen’s most lucrative products, and victory in patent infringement litigation has been crucial to its financial success. In this respect, the Epo story is similar to patent battles over small-molecule drugs. Another feature of the story is that Amgen started small and grew large because patents protected it from competition against pharmaceutical giants that were slow to appreciate the future value of recombinant DNA products. Amgen, Genentech, Genzyme and other first-generation biotechnology firms share this reliance on gene patents as part of their core business.
Polymerase Chain Reaction
The polymerase chain reaction (PCR) method was invented primarily by Kary Mullis at Cetus Corp., starting in 1983 (149, 179). Cetus patented the method (U.S. patents 4,683,202 and 4,683,195) (148, 151) and sold rights for most uses (except DNA forensics) to Hoffman--La Roche for $300 million, as Cetus merged with Chiron in 1991. The deal was complicated by a lawsuit in which Kodak tried to block the sale of PCR rights based on an earlier licensing agreement. PCR generated an estimated $2 billion in revenues for its rights holders before its initial patents began to expire in 2005 (79).
PCR was a technique for making copies of DNA segments quickly, with high fidelity, easily, and at relatively low cost. It spread like wildfire into molecular biological research and also found practical applications in diagnostics, DNA forensics, pharmaceutical and biotechnology R&D, and many other fields. The scientific community loved the method but pushed back on some of the ways Cetus tried to use its patent rights. Cetus initially tried to ensure future rights in discoveries made using PCR, but scientists and others objected noisily. The patents on the PCR method were linked to instrumentation for heating and cooling reaction mixtures in “thermocycler” instruments, a part of the process, and to an additional patent that Cetus secured on heat-stable DNA-replication enzymes that made the technique far easier to use (U.S. patent 4,889,818) (87). Most of the revenue streams came not from being able to directly monitor use of the method but from tying licenses to the reagents (e.g., Taq polymerase, developed from bacteria growing in the hot springs of Yellowstone National Park, Wyoming) and licenses on the thermocycler instruments.
PCR had a tortured legal history. In 1989, DuPont filed suit against Cetus contending its PCR patents did not meet patent criteria of novelty and nonobviousness. In response, the patent office initiated a re-examination of the patents. Prior publications of MIT’s Gobind Khorana were brought to the attention of the patent office, but the patent office reissued the patents after concluding that some key features of PCR, including exponential amplification, had not been disclosed in the prior art. A jury found for Cetus in February 1991 (150). As noted above, Kodak sued to block sale of PCR rights to Hoffman--La Roche in 1991 but lost.
Patent battles erupted on two more fronts. In 1992, Roche sued Promega Corp. over infringement of its patent for Taq polymerase (79, 105). Promega was selling the enzyme under a non-PCR license, and Roche accused it of inducing infringement. In 1995, Roche produced as evidence a list of scientists whose publications indicated use of PCR. Rather than suing the direct infringers at research institutions, Roche sued the firm selling them the enzyme without a PCR use license. This became a very complicated case. The initial judgement was that Cetus’s patent on the enzyme was invalid, but on appeal that was partially reversed (106) and remanded to the trial court. Judge Vaughn Walker reaffirmed invalidation of the Taq patent in May 2004 (107). By then a modified enzyme had become the main one used in PCR and the original PCR patents were on the verge of expiration. The other battle was about licensing thermocycler instruments for PCR. In June 1998, Roche sued MJ Research for selling instruments used for PCR without a license. Applied Biosystems (Applera) joined Roche in the case, which went to trial in 2004. A jury found infringement, and the judge doubled damages for willful infringement (18).
The upshot of the PCR patent story is that a very widely useful method was discovered in a small biotechnology company and it was patented. The method itself was hard to monitor for infringement. Most income attributable to the patents came either from large firms partnering to share rights to the invention, or from end product sales that embodied additional patents on an enzyme and the thermocycler instruments used in PCR. The patent rights were important in a business sense, and the technique spread widely through the scientific community, but only after initial resistance to Cetus’s efforts to secure reach-through rights to future discoveries led to Cetus's backing off. Once it acquired most PCR rights, Hoffmann--La Roche licensed its products in a way that enabled broad use. Patent litigation punctuated the story, and the PCR method patents withstood challenge, but a patent on the Taq polymerase did not. It is simply impossible to know whether patents helped or hindered the adoption and commercializaiton of PCR, but empirical studies do suggest that any impact on the advance of science attributable to patenting must be modest, because the pattern of scientific papers citing use of PCR is similar to other fundamental molecular biology methods of the time (79).
From ESTs to SNPs and the HapMap via Bermuda
A battle over patents was part of a larger war over how to conduct the Human Genome Project. The story began with a debate about how and what DNA to sequence as the Human Genome Project officially got underway in 1990: Start to sequence protein-coding regions (cDNAs derived from mRNAs) or map and sequence genomic DNA? Another debate was about whether to continue using Sanger and Maxam--Gilbert sequencing methods or to use the new automated sequencing machines. And how much sequencing, compared to characterization and mapping, should be funded?
Expressed Sequence Tags (ESTs)
J. Craig Venter ran a laboratory studying neurotransmitter receptor genes in the NIH intramural research program. His laboratory was an early adopter of the Applied Biosystems automated DNA sequencing instrument developed from the Caltech prototype. Venter initially proposed to sequence parts of the X chromosome, and then other gene-rich regions, such as the tip of chromosome 4 where the Huntington’s disease gene was known to reside, but the responsible mutations were yet to be discovered. Venter then shifted his main effort to sequencing short segments of protein coding regions as a quick way to tag genes using sequences unique to them. In June 1991, Mark Adams and colleagues described extracting 600 protein-coding sequences from human brain and determining their DNA sequence as expressed sequence tags (ESTs) (1; 211, pp. 126--38). A month later, in a Senate briefing, Venter publicly announced that the ESTs were the subject of a patent application (U.S. patent no. 07/716831) (213), filed one day before the June 1991 EST article was published. Controversy erupted when Science did a news feature on those patents in October 1991 (53, pp. 311--19; 187). The dispute was mainly about the politics of how to conduct the Human Genome Project, but one component of that argument centered on the EST patent applications, and the respective roles of public and private sectors (28, 69).
The NIH EST patent controversy galvanized the scientific community. The lawyer responsible for filing the NIH patent applications, Reid Adler, defended his action in Science beside a counterpoint article by Genentech patent lawyer Tom Kiley (5, 125). NIH director Bernadine Healy supported the patent application in the New England Journal of Medicine, arguing that she needed to keep the NIH’s commercialization options open (101). When Harold Varmus took the reins as NIH director, he brought with him a history of engagement with patent issues through the National Academy of Sciences. He commissioned patent scholars Rebecca Eisenberg and Robert Merges to give him advice about what to do with the NIH’s EST patent applications. In a closely reasoned, 52-page document, they urged Varmus to pursue the patents only if he judged they would advance commercialization without hindering science, and they laid out arguments why prospects of commercialization were dim but opportunities for impeding science were real (72). Varmus took their advice.
The EST patent controversy quieted down for several years when NIH abandoned its EST patent applications in 1994 but then roared back to life three years later when the U.S. patent office signaled it was about to grant patents on ESTs. The announcement came at a symposium on gene patents in February 1997 (37). The patents being examined were not from the NIH but from companies that had incorporated DNA sequencing of gene fragments into their business strategies. John Doll of the USPTO published an article in Science explaining the rationale (65). The scientific community was having none of it, however. Varmus sent a letter from the NIH to the patent commissioner arguing for strong evidence of utility in granting DNA sequence patents (210), and Bruce Alberts sent a letter as president of the National Academy of Sciences (8). Concern over ESTs converged with the 1997 CAFC decision in UC v. Lilly (184), which also raised concerns about the thresholds for utility and specific written description requirements for sequence-based patents. The result was new examination guidelines---proposed formally in 1999 and finally promulgated in the Federal Register in early 2001---that required “specific, substantial, and credible” evidence of utility (61) and reinforced the written description standard (62). In October 1998, the USPTO did issue at least one patent, U.S. patent no. 5,817,479 (23), on genes encoding multiple kinase proteins , and that action attracted some notice (188) but no major controversy ensued---no doubt because the patents were not enforced against researchers.
The EST controversy, commercial genomics, and data-sharing practices
The EST patent controversy had several consequences, many of which were salutary but utterly unpredictable and inadvertent. One consequence was that it elicited business interest in genomics. Randall Scott worked at Incyte, a small biotech start-up that was mainly doing contract research for Genentech. The EST patent controversy drew his attention to the potential of sequencing protein-coding regions and patenting all or parts of genes as a business strategy, and Incyte revamped its R&D along those lines. Wallace Steinberg, an angel investor, likewise learned about Craig Venter because of the EST patent controversy. He approached Venter and eventually lured him into the private sector to found a nonprofit research organization, The Institute for Genomic Research (TIGR), which would focus on sequencing but would vest patent rights in a for-profit company, Human Genome Sciences (HGS). Both Incyte’s and HGS’s strategies centered on sequencing protein-coding DNA, filing patents, and either developing products or licensing rights. The EST controversy led directly to these businesses beginning to focus on genomics.
The EST patent controversy also made clear that the organizations funding the Human Genome Project needed to think explicitly about when and how to share DNA sequence data and other information, and what to do about patents on research funded through the Human Genome Project. The Caenorhabditis elegans and yeast genomics communities became activated in a movement to preserve freedom to operate in genomics, and their model of how to conduct science spilled over to the Human Genome Project (15, 16). Funding agencies and scientists realized that they needed systematic policies to cultivate a scientific commons, lest they lose control of their science to those wielding patents. Four examples of such collective action illustrate how norms of open science were put in place: (a) the public domain EST sequencing projects funded by Merck and the DOE, (b) the Bermuda Rules of sharing sequence data rapidly, (c) the SNP Consortium, and (d) the HapMap project.
Merck’s public domain cDNA sequencing effort
As Human Genome Sciences, Incyte, and other genomic start-up companies began to form in 1991--1993,1 Alan Williamson at Merck worried about proliferation of patents on DNA fragments, full-length protein-coding genes, and other inputs to pharmaceutical R&D. Merck decided to fund Washington University at Saint Louis, one of the largest DNA sequencing centers, to identify and sequence protein-coding regions and deposit them into the public domain where they could not be patented and, moreover, would block others from patenting. Merck funded this initiative on the rationale that it would accelerate science, retain freedom to operate, build good will among scientists with whom Merck had many collaborative R&D projects, and at least partially block small start-ups from controlling crucial gene patents that they could use to extract revenues from the likes of Merck (222, 223). Three of these four benefits would also redound to competitor pharmaceutical and biotechnology firms. The elements distinctive to Merck were branding, good will, and close, informal working relationships with cutting-edge genome scientists. Merck funded this effort through a nonprofit arm, which meant it could have no special access to the resulting data, but that openness fit the purpose of the effort. The resulting database became a scientific resource for not only Merck and academic researchers but also biotechnology and pharmaceutical R&D (88). It cost Merck several million dollars to fund the sequencing, but putting the data into the public domain was intended to forestall future costs should Merck have to negotiate with Incyte, Human Genome Sciences, a university, or some other patent holder every time it used a gene. Eisenberg remarked on the anomalous topsy-turvy world in which a pharmaceutical firm would fund open science that the NIH did not fund (68, p. 561). Yet it did make sense.
Bermuda Rules
A fear of private entities sequencing and patenting genes rapidly drove the Human Genome Project funders to vigorously protect the public domain as DNA sequencing began to take a more prominent place in the Human Genome Project in the mid-1990s. The Wellcome Trust spearheaded a 1996 meeting in Bermuda to forge principles among the high-throughput sequencing centers that were beginning to produce data rapidly. The meeting participants agreed to make sequence data available within a day once a contiguous stretch of 1,000 nucleotides had been assembled. This policy had two principal rationales: (a) it reduced concerns among small laboratory users of the data that highly capitalized centers would examine the juicy bits of the genome first, leaving only the crumbs,for smaller enterprises, and (b) it effectively prevented a patent logjam on genes and other sequences because public release of data would preclude patenting of the sequences. It would not necessarily block patents on genes and other sequences if someone found a function and did something inventive, new, and useful, but it would prevent the kind of sequence-based patents such as the ESTs that the NIH had tried to patent, and sequence-based patents for which Human Genome Sciences, Incyte, Ohtsuka, and other firms were known to be applying.
The SNP Consortium
Even as DNA sequencing was ramping up, microarray technologies broke onto the scene. Patrick Brown at Stanford, Edwin Southern at Edinburgh and then Oxford, a Stanford spin-out company Affymetrix, and other groups began to develop technologies to hybridize many thousands (eventually millions) of sequences to DNA from samples of many kinds (individual genotypes, gene expression profiles, tumor samples, etc.). At the same time, techniques for identifying single nucleotide DNA sequence differences, or single nucleotide polymorphisms (SNPs), advanced rapidly. Several companies signaled that they intended to identify and patent such SNPs, and again the prospect of a thicket of patent rights atomized among innumerable patent holders, or even worse, held by a single firm, led to an agreement to form a public-private partnership, the SNP Consortium, to discover and characterize SNPs and to ensure they remained in the public domain. Because private players were already in the race but their activities could not be reliably monitored without their cooperation, the Consortium devised an elaborate IP process.
Many SNPs were systematically discovered at high-throughput centers, which filed provisional patent applications to establish priority dates and standing as inventors in case others later filed patent applications on the same SNPs. This freed the patent applicants to share data without losing rights as inventors. They kept data secret until the SNPs were characterized and mapped to the chromosomes, at which point they could be either converted to statutory invention registrations (which provided no exclusivity, but did prevent others from getting patents) or simply abandoned, thus releasing the data into the public domain (214). This was a highly creative, but laborious and relatively expensive, way to ensure freedom to use SNPs in the future.
International HapMap Rules
A final stratagem of note was the data-sharing policy associated with the human Haplotype Map, or HapMap. Haplotypes are clusters of DNA markers that tend to be inherited together, because DNA exchange during meiotic cell division is relatively infrequent in each generation, and so markers that are close to one another will tend to be inherited together as blocks. The HapMap was an effort to identify enough markers throughout the human genome to be able to trace the inheritance of DNA from chromosomal regions. This required sampling individuals in many populations to look for DNA variants to be used as markers. The HapMap project was inherently large-scale and collective. It was funded by many agencies and organizations that had been involved in the Human Genome Project, augmented by new partners (119).2 The funding for research and use of data required agreement to a set of data-sharing rules. The rules included not seeking patents on haplotypes or SNPs and not sharing data with others who did not agree to the license. This preserved freedom to operate, but there was some ambiguity about what kinds of patents might be permitted. The initial rules also precluded any patent incentives that might be needed to develop commercial uses of the SNPs and haplotypes (74; 180, pp. 215--216). The strict HapMap rules were relaxed to permit use and selective patenting, once there were ample SNPs in the public domain, and as realization dawned that private investment in commercialization might be desirable in some cases, and might be easier to attract with patent incentives.
Celera and the Great Genome Race of 2000
In May 1998, the Human Genome Project became a race of sorts, between the public effort funded by governments and nonprofit philanthropies and a new start-up company that took the name Celera later that year. Many events led to this emergence of a privately funded, corporate large-scale sequencing effort, but two are of particular note. One was success in mapping full-genome sequences of bacteria and other whole organisms. The publication of the Hemophilus influenzae genome in June 1995 was a watershed event. It was immediately recognized as a powerful tool for studying this bacterial pathogen, and also a demonstration of what was termed the whole-genome shotgun sequencing strategy. Whole-genome shotgun sequencing started by generating masses of DNA sequence data and relying on computer assembly of the sequence from overlapping sequence reads. This contrasted with a map first strategy that turned to DNA sequencing only after intermediate steps of assembling genomic maps and aligning DNA fragments.
A second generation of automated DNA sequencing instrumentation was another contributing factor. ABI’s workhorse sequencing instrument, the Prism 377, dominated the DNA sequencing market by 1997 but was being threatened by a new competitor MegaBACE, a capillary gel sequencer from Silicon Valley start-up Molecular Dynamics (then having just been acquired by Amersham Pharmacia). Capillary gels used less DNA, generated data far faster, and collected more data per run. The idea of using 200 of ABI’s planned new instruments to sequence the entire genome in three years grew out of a November 1997 board meeting (194, pp. 64--67). ABI’s Mike Hunkapiller became the person who introduced the idea to Craig Venter at TIGR. Nicholas Wade was given a New York Times exclusive to announce the intention to sequence the human genome in a for-profit corporation (216). Venter resigned from TIGR to head up the new company, which proceeded to use the new ABI capillary-gel sequencing instruments to first sequence the Drosophila genome and then the human genome, in parallel to the publicly funded effort.
From the initial announcement about forming a new company to sequence the genome by whole-genome shotgun sequencing in May 1998, through the June 2000 announcement of a draft human genomic sequence that had Craig Venter and Francis Collins flanking President Bill Clinton in the White House, and until the February 2001 twin publication of draft genomic sequences in Science and Nature (131, 212), the story was framed as a race, despite repeated denials by those engaged in it---and that race was the biggest story in all of science. It was often reported as a corporation bent on patenting the human genome pitted against the publicly funded effort that eschewed patents, but there was little truth on either side of this equation. The high-throughput sequencing centers were abiding by the Bermuda Rules, which precluded direct patenting, but many other gene hunts were going on at the same time in academe, and universities were seeking DNA patents. Celera did file patent applications, but Craig Venter acquired a grand total of 15 U.S. patents, most of them from his period at TIGR, and most of those abandoned after four years (59; web search by R. Cook-Deegan performed on December 4, 2009).
The efforts at Incyte, Human Genome Sciences, and other companies to identify, sequence, and patent genes were much more relevant to concerns about patent impediments to genomic sequence data than Celera’s sequencing program. The inchoate fear of the genome being locked up and patented was a common topic at meetings of genome scientists and it dramatically increased attention to norms of sharing data, which accelerated progress. In the private sector, the widespread availability of genomic sequence data also no doubt reined in the proprietary instincts of the corporate effort. Efforts to publish the results of Celera’s sequencing program were sources of tremendous conflict within Celera, just as they had been between TIGR and its parent company Human Genome Sciences (194, 211). A robust publicly funded source of DNA sequence data both goaded further publication of privately generated sequence data to verify claims being made about it and ensured that trade secrecy would not be an option to protect genomic sequence information. The most reliable protection for proprietary genomic data became patents, and the highly public race to sequence the genome was no doubt a stimulus to file DNA patent applications. The flood of data also raised the bar on what an inventor would need to show to prove utility, nonobviousness, and novelty.
Patents and Instruments used in Genomics
Patents have played an important role in the development of several instruments used in genetics and genomics research and its applications. The DNA sequencing instruments used to carry out the Human Genome Project were based on the unpatented Sanger sequencing method. The instruments to automate DNA sequencing, however, entailed considerable engineering, such as finding four-color fluorescent labels for DNA molecules, optical detection methods, electrophoretic separation techniques, and software algorithms to interpret raw data into DNA sequence information. Many DNA sequencing instruments were developed, but the dominant technology for the Human Genome Project grew from an instrument developed at Caltech as a prototype and manufactured by Applied Biosystems (ABI, now part of Life Technologies) (54, 110, 196). This was a classic university spin-off story, with Caltech research giving rise to a patented method that was initially licensed exclusively to ABI when it was a nascent biotech start-up firm. The story was not without conflict, however. There was initial skirmishing for market dominance among instrument developers ABI, DuPont, Amersham, and EG&G, and Japanese firms. Henry Huang, who left Caltech where he had been involved in early DNA sequencing automation efforts, sued over being excluded as an inventor on four patents. In February 2004, he lost decisively in court (104).
The early history of Affymetrix is likewise a university spin-off story, centered on adapting lithography techniques used to make semiconductor chips for use as DNA microarrays. Patents on the methods were key to the story, as were federal grants to foster commercial development of the nascent technology. This Silicon Valley story involved Stanford, the Advanced Technology Program, and a succession of start-ups that gave rise to Affymetrix, Perlegen, 23andMe, and other genomics companies that took root in the fecund genomic soil of the San Francisco Bay Area (132).
David Walt’s insights into microbeads and fiberoptic detection methods at Tufts University gave rise to several patented inventions that helped spawn Illumina. Illumina began as a microarray technology company and has since become a leader in DNA sequencing technology, in part through its acquisition of Solexa. Illumina’s early history involved Tufts patents exclusively licensed via a Massachusetts multi-university incubator consortium, before Tufts developed its own technology licensing office (172). Both Affymetrix and Illumina are classic Bayh--Dole stories of federally funded university research giving rise to patented technologies licensed exclusively to start-up firms that produced instruments valuable for research. The initial market for their instruments was a combination of academic research laboratories and R&D laboratories of biotechnology and pharmaceutical companies. Affymetrix and Illumina then cultivated commerical applications such as diagnostics and forensics.
Litigation has been common among genomic instrumentation firms, and between them and larger established firms. Some of that legal conflict centers on specific patents. Oxford Gene Technologies, which had exclusive licenses to microarray patents of Edwin Southern, won U.S. and U.K. lawsuits against Affymetrix in 2000 (6, 51, 168, 169). The firms settled while a separate trial challenging the validity of Oxford’s patent claims was underway. Affymetrix settled disputes with HySeq and with Incyte in 2001 (134, 200). Affymetrix and Illumina have been engaged in a long-standing series of lawsuits against one another over microarray technology, including an ongoing suit initiated in May 2009. Illumina and ABI (now Life Technologies) have several ongoing infringement lawsuits against one another over DNA sequencing patents, filed from December 2006 to October 2009 (80, 82, 83, 117, 133).
Patent litigation is rife, and patents in DNA sequencing instruments and microarray technologies have been hotly contested. Little of this conflict has spilled over from the business sections of the news media onto the front page or into the scientific journals, however, in stark contrast to the extremely public disputes over DNA diagnostics, which have ironically yielded relatively little actual litigation. The reason for litigation is that in the instrumentation business, patents have become tools for extracting revenue streams from competitors, and the payoff for both sides is sufficient to warrant going to battle in court. Affymetrix, Illumina, Life Technologies, and other firms remain active in their markets despite wins and losses in court; none has entirely withdrawn from the market. Moreover, most of the lawsuits are between competitor firms battling for markets, a contrast with the notification or cease and desist letters in genetic diagnostics, which have been the most public and most controversial when directed against university genetic testing services, and which have generally led to withdrawal from the market. That is, in instruments, patents have been used in their accustomed role of solving disputes among for-profit business firms competing for future profits. Patent suits have generally not completely eliminated a competitor. Moreover, patent litigation has generally resolved who makes money from selling instruments, not who controls access to a life-saving technology. The fights have been about money, not about clinical patient access or the progress of research and media interest has therefore mainly been confined to the business pages.
The Anticommons Debate
Just two weeks before the formation of Celera was announced, Science published one of the seminal articles about DNA patenting in the annals of scholarship. Michael Heller and Rebecca Eisenberg described how an “anticommons” might form with the patenting of many inventions far upstream from final application (103). The idea originated from Heller’s broader theory derived from studying Russia’s transition from communism to a market economy (102) and got theoretical support from Nobel economist James Buchanan and Yong Yoon (35). If intellectual property were too fragemented, it could make it difficult to collect all the pieces needed to move forward toward practical application. This was a different concern from individual patents that might block others, because genes as finite objects of nature cannot be worked around if patented. It was also separate from concerns about broad patent scope, in which claims exceeded what an inventor had actually discovered, reduced to practice, and described in a patent and thus over-reached, fencing in more intellectual territory than patent law should in theory allow.
The anticommons idea caught fire and was widely discussed among scholars and scientists concerned about DNA patents. It generated a mini-literature. Murray and Stern found a mild inhibitory effect of patenting on subsequent publications in Nature Biotechnology (152). Another analysis used 2,647 patented sequences, a subset of the Jensen & Murray data set cited above, and looked for effects on future citations in the scientific literature after a patent was granted. Huang and Murray found a 5% decrement in such citations (17% by a less stringent metric). The drop in citations was larger with cancer compared to noncancer genes, disease-associated compared to non-disease-associated genes, and for genes listed in Mendelian Inheritance in Man compared to those not listed, leading the authors to conclude, “The more immediately useful and relevant and commercializable the patented (genetic) knowledge, the more negative the impact of gene patent grant on subsequent published citations” (113).
Walsh and Cohen surveyed scientists and found that few checked to see if they were infringing patents and only a few percent reported an effect of patents on their scientific projects. To the degree that there was friction in research, it was more attributable to material transfer agreements of tangible research reagents (219, 220). It was hard to find empirical evidence of substantial slowing in the progress of genomics (39). And a scholarly literature accumulated about theoretical reasons to be skeptical of a big anticommons effect (e.g., 3, 4, 193). David Adelman, for example, observed that “research opportunities far exceed the capacities of the scientific community. It is this basic dynamic that makes biotech science, in important respects, an effectively unbounded, uncongested common resource” (2, p. 987).
The argument is not over, however. Scholars have mainly looked at research for evidence of anticommons effects. Research is indeed a market for genomic research tools, but it is not the place where difficulties in assembling legal rights to enter commercial markets would have their biggest effects. Who wants to sue scientists for doing research? Patent holders can benefit from research using their inventions. What would the damages be? Stronger anticommons effects would be expected farther downstream, in foregone investment in innovation that required patented inventions as an input. Eisenberg re-visited the evidence and concluded that there were few anticommons effects in most research (with the exception of transgenic animals) but that, in diagnostics, there were some indications of difficulty accumulating rights and impacts on market entry. Moreover, the situation might be metastable, because the main reason that academic scientists have not encountered problems with patents is that they ignore them. She argues for adjusting “the patent law to reflect the norm rather than relying on noncompliance and nonenforcement under the current law” (71, p. 1097).
In this framework, the effects of patent thickets and anticommons effects on innovation and access are more likely to be found in clinical genetic testing than in research. We now return to genetic testing to examine the evidence in more detail.
BRCA: An Outlier
The level of public furor over genetic diagnostic testing has only recently led to patent litigation. As noted above, public media and policy reports have barely noticed the frequent litigation in instruments and therapeutics, but they have devoted significant attention to genetic diagnostic controversies and potential impediments to research, largely because of the direct patient impact and compelling personal stories of prospective users, and sole-provider business models that confer controversial national monopolies among genetic testing firms. BRCA testing for genetic susceptibility to breast and ovarian cancers has been particularly conspicuous. BRCA was cited repeatedly in passage of legislation to create a compulsory licensing authority and to expand research use exemptions in Europe (207, 208, 209), was by far the most cited gene patent controversy in media accounts and policy reports (38, 39), was the subject of two of five cases of patent litigation in diagnostics reviewed by Holman (108, 109), and is now the subject of the first genetic diagnostic case to go before a U.S. judge. It led to a starling ruling by Judge Robert Sweet of the Manhattan federal district court in New York, discussed below, that invalidated the broadest method and sequence-based claims of patents on BRCA1 and BRCA2 in a 158-page ruling made public on March 29, 2010 (21A).
What is so special about BRCA that it elicits such a vigorous public debate? Several features are distinctive to the case. BRCA genes were the subject of a particularly intense race to discover the genes associated with cancer risk, and then to patent them. The race was set off in fall 1990, when Mary-Claire King found genetic linkage between cancer risk and markers on chromosome 17 in families with many cases of breast and ovarian cancer, consistent with dominant Mendelian inheritance in those families (98). Such familial risk accounts for only 5--10% of all breast cancers but a significantly higher fraction of those occurring before age 50)(98). Mutations in the gene that became known as BRCA1 were identified in 1994, and mutations in a second gene on chromosome 13 (BRCA2) were found the following year. A team at the University of Utah and Myriad Genetics crossed the finish line just before other competitors for BRCA1, and the winner of the BRCA2 race remains controversial to this day (55, 94).
The patent story associated with these races is complicated. The following account is summarized from Gold & Carbone (94) and Cook-Deegan et al. (55). After initially being left off the Myriad/Utah patents, two scientists from the National Institute of Environmental Health Sciences (part of the NIH) were added as coinventors. The NIH then assigned administration of the patents to Utah, which licensed its BRCA patent estate exclusively to Myriad. The first patent granted on BRCA1 was a U.S. patent held by Oncormed, which also licensed patents resulting from Mary-Claire King’s work at the University of California. Myriad got its own patents on BRCA1. Oncormed sued Myriad (165) and Myriad countersued (153). Myriad also sued the University of Pennsylvania (154), which was already offering BRCA genetic testing when Myriad entered the market in. Indeed, based on information available from a survey of laboratory directors by Mildred Cho and colleagues (43), Penn was not the only laboratory to beat Myriad to market with a BRCA test. Penn and eight other laboratories, most of them university clinical testing services, were also offering BRCA testing and ceased offering it in response to Myriad’s lawsuits and cease and desist enforcement letters (43, Tbl. 2).
Penn bore the brunt of another related conflict in 1999, when it offered to do genetic testing for National Cancer Institute (NCI) clinical trials and other federally funded clinical research. Myriad raised objections, and NCI director Richard Klausner signed a memorandum of understanding with Myriad that ensured Myriad would do most BRCA testing for such clinical research. Other laboratories could do their own BRCA testing only if it were performed for patients at the institution itself (i.e., Penn could do BRCA testing for Penn but not as a service for other research centers) and if results of testing were not returned to research participants (139). In return, Myriad offered a deep discount for BRCA testing done for academic research.
Myriad thus established itself as sole provider of BRCA testing in the United States by enforcing or threatening to enforce its patents. This sole provider business model has not worked in any other jurisdiction, even when patents similar in scope to U.S. patents have been granted (94, 173).
In Canada, Myriad licensed its rights to MDS, a private firm, which with Industry Canada encouraged Myriad to rattle its sabre and threaten patent enforcement against provincial health authorities. In 2001, Myriad sent cease and desist letters to health ministries in four provinces: Quebec, Ontario, British Columbia, and Alberta. Tony Clement, Ontario’s health minister and thus head of MDS/Myriad’s single largest potential customer in Canada, conferred with Ontario’s prime minister, Mike Harris, and they decided to push back. Myriad called for a meeting with Clement and brought two threatening letters to the meeting---one from the U.S. Ambassador to Canada alluding to trade sanctions and another from Senator Orin Hatch of Utah (Myriad’s home state), indicating he was “watching” the situation closely and had alerted the U.S. Trade Representative. The Biotechnology Industry Organization also threatened to cancel its planned annual meeting in Toronto (94, pp. S51—S53). This hamfisted overreach backfired, and Clement called Myriad’s bluff. Myriad’s alternative now was to sue its largest Canadian customer, the Ontario provincial health system. Indeed, the politics were easy. Any Canadian health minister seen to knuckle under and relinquish control of testing Canadians for breast cancer to a U.S. corporation was doomed to a bloodletting in the Canadian press. Instead, it was Myriad that lost its main opportunity to enter the Canadian market, as it has never sued to enforce its patents, and over time the provincial health programs have resumed unlicensed BRCA testing.
The situation is similar in the United Kingdom, where Myriad has fairly strong patents, but the National Health System has largely ignored them (173). In Australia, Myriad was itself being threatened by Genetic Technologies, Ltd. (GTG) for use of its patents claiming use of DNA sequences between genes. As part of an agreement, GTG became Myriad’s licensee for testing in Australia and New Zealand but decided not to enforce patents against the provincial health authorities there, instead announcing it was allowing them to do BRCA testing as a “gift” to the people of Australia. As GTG became financially stressed in 2008, it announced that it had changed its mind. This provoked a tremendous backlash in the Australian press as well as an investigation by the Commmittee on Community Affairs of the Australian Senate, which is scheduled to produce a report in summer 2010 specifically considering whether DNA sequence patents should be permitted in Australia (55; R. Cook-Deegan, personal communication with Elton Humphrey, Committee Secretary, Committee on Community Affairs, Australian Senate). A lawsuit against Myriad and GTG was filed in Australia in May 2010 (36A), and legislative proposals for a diagnostic use exemption and banning gene patents entirely have been proposed as statutory solutions in Australian law.
Patents on BRCA1 and BRCA2 in Europe were challenged by a coalition of organizations including academic research and clinical institutions and health professional groups, using the opposition authority in the European Patent Office (94, p. S45). Two BRCA1 patents whose claims were revoked in 2004 while the opposition proceedings were under way have been partially restored, but the scope of their claims is to mutations that Myriad had demonstrated at the time of patent application (55, Feb. 2010 update).
The fight over BRCA patents has now returned home to the United States. In May 2009, the Association of Molecular Pathologists and a group of individuals and health professional organizations sponsored by the American Civil Liberties Union filed suit against Myriad Genetics, Utah, and the U.S. Patent and Trademark Office in the Southern District Federal Court in New York (20). Judge Robert Sweet released an 88-page opinion in November 2009 that indicated his intention to hear the case (21), and his ruling of March 29 invalidated all the contested claims in patents held by Myriad (21A). The lawyers on the ACLU side include Christopher Hansen, whose main interests are first amendment rights, and Dan Ravicher, director of the Public Patent Foundation, a former corporate patent lawyer now affiliated with Cardozo Law School. In their public statements, they have made plain their aspirations to directly challenge the legitimacy of all “gene patents,” and to appeal the case up to the U.S. Supreme Court if possible (7, 147). The plaintiffs include two of the clinicians whom Myriad sued a decade earlier at the University of Pennsylvania and several individual women who want genetic testing and claim Myriad’s policies and practices prevent them from getting their test without risking infringement. It is clear that the purpose in bringing the suit is to challenge Myriad directly as an exemplar of gene patent practice that uses patent rights to enforce a sole provider service model, and to take the case as far up the chain of federal courts as possible as a precedent-setting case.
In choosing BRCA and suing Myriad to make its case, the ACLU selected a case that was already highly conspicuous, built on Myriad’s overwhelmingly negative public image based on media coverage, and Myriad’s past record of having been the most litigious genetic testing company (e.g., the only one to have sued a university) and having shut down nine testing services.
The case concerns a life-threatening cancer associated with highly organized breast cancer constituency organizations (several of which support the ACLU side and none of which support Myriad; and many are neutral). Myriad also finds itself defending some especially broad U.S. patent claims.
Myriad does have some conditions in its favor. The company is generally regarded as offering good clinical services with accurate reports and good turnaround time. Its enforcement actions took place over a decade ago, and some of the strong antipathy those actions generated has dissipated. Myriad's unit costs for sequencing the two large BRCA1 and BRCA2 genes are actually slightly lower than what it and other laboratories charge for testing colorectal cancer susceptibility genes (55), and considerably lower than what Athena Diagnostics charges for sequencing smaller genes for other conditions and what PGxHealth charges for Long-QT genetic testing (14, 175, 195). Myriad began to offer augmented testing for chromosomal rearrangements in 2006 when problems with its sequence-based testing became public. And it is defending its patent rights in the United States, one of the most patent-friendly jurisdictions in the world.
Judge Sweet’s decision is being appealed to the Court of Appeals for the Federal Circuit, where many patent lawyers expect it to be reversed at least in part. If so, that could lead, in turn, to an appeal to the Supreme Court. At this point, however, it is mere speculation to guess either the outcome of this particular case or its implications for gene patents in diagnostics in the United States. But the first round was a shocker, particularly to practitioners of patent law.
The federal district court ruling has injected a new level of uncertainty about gene patents, particularly as used in diagnostics. This was not only the first diagnostic case to progress so far in the U.S. court system, but it is also unusual in that the plaintiffs were not commercial rivals but potential users of the test: patients at risk who wanted to get tested or physicians or health professional organizations representing doctors who order genetic tests for their patients. This unusual feature of the case may explain the befuddlement of the patent lawyers generally accustomed to patent litigation that pits one commercial firm against another rather than potential customers suing a service provider.
The case has rekindled public attention to the BRCA patenting controversy and the practices of firms that use patent rights to create sole-source genetic testing business models. Myriad Genetics will have to find a new business model in any event, since its BRCA patents will begin to expire in 2014 and 2015.
Genetic Testing Beyond BRCA
BRCA testing is the most public debate and the most litigious case in clinical genetic testing, but genetic tests for many other conditions have also been drawn into a debate about patents. The European Society of Human Genetics published recommendations in 2008, noting that patents are often beneficial but observing that the effects of patents on genetic diagnostics were “intrinsically different from patenting of methods, tools, and technologies.” A task force recommended narrowing patent scope of claims that could affect diagnostic uses, establishing a reporting system for problems, and exploring patent pools and cross-licensing solutions to free up access to the requisite technologies (24). As noted earlier, the SACGHS also prepared a report on the topic in 2010 (190A).
Two bodies of scholarship have begun to enrich the debate about patents and genetic testing. Van Overwalle and colleagues identified genetic tests for the 22 conditions most commonly the subject of genetic testing in Europe. They identified 250 relevant patents (in 72 “families,” i.e., groups of patents related to the same underlying invention in different jurisdictions) and analyzed the patent claims (116). Patents were associated with 19 of the 22 diseases, and there was at least one blocking claim in at least one patent for 15 (68%) conditions. Only about 15% of the claims analyzed were deemed “blocking,” and among those claiming “genes,” this designation was given to only 3%. Method claims were more apt to prove difficult to work around than other kinds of claims. However, counting the percentage of claims that are blocking does not indicate the extent to which exclusive rights affect clinical testing. The better indicator of how patent rights might affect clinical use is the percentage of the conditions for which at least one patent is blocking (i.e., 68%).
Another source of empirical data about patents and clinical access to genetic testing comes from case studies done for SACGHS in 2007 and 2008. Eight case studies addressed ten clinical conditions selected by the committee to be informative (14, 40–42, 48, 55, 175, 195). Table 1 summarizes the findings from those case studies.
Table 1.
Patenting and Licensing for Ten Conditions with Mendelian Inheritance1[**AU: See this copy below as table footnote 1**]
| Medical condition (test providers) | Gene(s) associated | Patent/licensing status |
|---|---|---|
|
Inherited risk of breast and ovarian cancer (Myriad dominant in United States) |
BRCA1, BRCA2 | Patents owned by universities and Myriad Genetics. Exclusively licensed to Myriad in United States. |
|
Inherited risk of colorectal cancer (Myriad and others) |
APC, MYH (FAP and attenuated FAP) MLH1, MSH2, MSH6 (Lynch Syndrome) |
University patents nonexclusively licensed |
|
Tay--Sachs disease (various providers) |
HEXA (enzyme function usually tested) |
HEXA gene patent owned by National Institutes of Health; not licensed |
|
Canavan disease (various providers) |
ASPA | Miami Children’s Hospital Research Institute owns patent; initial restrictive licensing; confidential settlement |
|
Cystic fibrosis (various providers) |
CFTR | University patents nonexclusively licensed |
|
Alzheimer’s disease (Athena Diagnostics dominant in United States) |
Early onset: APP, PSEN1, PSEN2 |
PSEN2 university patent exclusively licensed to Athena; PSEN1 and APOE university method patents exclusively licensed to Athena; APP unpatented for diagnosis |
| Late onset: APOE | ||
|
Spinocerebellar ataxia (Athena Diagnostics dominant in United States) |
30+ autosomal dominant genes (also recessive and X-linked but not studied) |
SCA1, 2, 3, 6, 7 & 8 exclusively licensed to Athena; mostly university owned; SCA-10 university patent nonexclusively licensed to Athena; Athena owns patent for Aprataxin; others unpatented to date |
|
Hemochromatosis (various providers using Bio-Rad tests) |
HFE (most common) | Patents owned initially by Mercator Genetics; current owner Bio-Rad Ltd.; initially exclusive licensing; now nonexclusively licensed |
|
Hearing loss (Athena Diagnostics main provider but several others; sublicense to Pediatrix) |
100+ genes; many mutations Connexin 26, 30, MTRNR1, MTTS1, SLC26A4 commonly tested |
Just two out of five of the most commonly tested genes have patents owned by nonprofits; exclusively licensed to Athena; most other patents university owned |
|
Long-QT syndrome (PGxHealth dominant in United States, with different mutations licensed to BioReference Labs) |
11+ genes | University patents on several mutations and genes exclusively licensed PGxHealth, and on other genes and mutations to GeneDx; both PGxHealth and GeneDx now offer test for alleles in 10+ genes |
Summary of findings from eight case studies prepared for a task force of the Secretary’s Advisory Committee on Genetics, Health, and Society, U.S. Department of Health and Human Services http://journals.lww.com/geneticsinmedicine/toc/2010/04001.
Distinctive Features of the Genetic Testing Market
One distinctive feature of genetic testing arises in how clinical genetic testing takes place. Exclusive rights to just one blocking claim for any gene associated with a given condition can discourage market entry by others and can often effectively secure market control for the exclusive licensee of one or a few key patents. This is because the purpose of sending a sample is often to identify which mutation in which gene might be responsible for clinical findings—the gene or allele cannot be known in advance, and so which patents might be infringed cannot be entirely predicted in advance. This is because one clinical syndrome might require testing for many genes and specific mutations covered by different patents. A laboratory offering testing services for that condition cannot know in advance which mutation will be found, and often not even which gene. Anyone testing for the condition will need rights to test for any variant likely to be tested, or risk infringement liability. If any firm holds exclusive rights to any method or sequence that might turn up in some patients, and if others lack countervailing exclusive rights, then the lone exclusive licensee can in effect secure the entire market as the only laboratory that can test for all variants. It can send enforcement letters to any laboratory testing for the entire condition because testing laboratories cannot know up front if they might in fact detect a DNA variant covered by a patent, even if most of the time their tests would not infringe. Thus a single blocking patent on a normal gene or any common disease-associated variant can be sufficient, if exclusively licensed to just one provider, to limit testing by other laboratories for that clinical condition.
This is how Athena Diagnostics became the sole provider of genetic testing for many neurological and endocrine conditions (including muscular dystrophies, Alzheimer’s disease, hereditary deafness, spinocerebellar ataxias, and other conditions). Many, indeed often most, mutations associated with the tested conditions are not patented---and for hearing loss, ataxias, and several cancers, there are many disease-associated genes for which there are no blocking patents---but having exclusive rights to one common disease-associated testing patent nonetheless can create monopoly power. Over two-thirds of the patents exclusively licensed to Athena are from universities or academic research institutions. Under this business model of exclusively licensing patents, mainly from academic centers, Athena had the most exclusive rights studied by van Overwalle et al. (116) and was responsible for most of the enforcement that led laboratories to withdraw from genetic testing in the 2003 survey of genetic testing laboratories by Cho et al. (43).
Similarly, PGxHealth, a subsidiary of Clinical Data, Inc., controlled the market for all genetic testing of long-QT syndrome until 2009, by having exclusive rights to patents on just five of many genes associated with the syndrome. When another firm, GeneDx (a subsidiary of BioReference Laboratories, Inc.) secured exclusive rights on other variants, however, the monopoly was broken, and both laboratories now offer genetic testing for at least ten different genes associated with long-QT syndrome (14, see November 2009 update). This situation makes clear the potential for mutual-blocking situations. In this case GeneDx’s acquisition of countervailing exclusive patent rights to some long-QT gene patents changed a monopoly into a duopoly for long-QT genetic testing in the United States. In other cases, it could instead require a more complicated cross-licensing scheme, litigation, or other solution if there are multiple blocking rights. Such patent logjams have occurred historically with sewing machines (144), aircraft manufacture, and radio broadcast. The process for resolving such patent thickets can be slow and conflict-ridden, but there are remedies for the kind of patent thickets that could arise in genetic diagnostics. None of the analogies is exact, but scholars are already contemplating how to solve such problems (e.g., see Reference 208 for several essays in a collection edited by G. van Overwalle, Gene Patents and Collaborative Licensing Models).
One additional distinctive feature of clinical genetic testing is that the targets of enforcement for patents exclusively licensed to testing firms include university laboratory services. This is quite different from a biotechnology firm blocking a competitor firm from entering the market with a rival drug or vaccine. There are several reasons that several academic health centers have genetic testing services. One is that they do research on genetic conditions and require the technical capacity to analyze DNA. Clinical testing requires certification under the Clinical Laboratories Improvement Amendments (CLIA), under federal jurisdiction of the Centers for Medicare and Medicaid Services.
Universities run many genetic tests, often because one research group began studying a condition, more patients with that condition returned for evaluation, and the laboratory was then required to initiate CLIA-certified clinical testing. Once a laboratory has certification for one genetic test, adding another genetic test means changing just the particular DNA sequences used, not developing an entirely new method or instrument. CLIA certification is given by laboratory, not by specific test. For this reason, it is a relatively small step for any laboratory already offering a genetic test to add another that uses similar methods but different DNA sequences. The gap from publication of a sequence associated with disease to testing for that sequence is short.
For a start-up firm or new laboratory, however, building the capacity to develop that same test from scratch can be expensive and entails getting an entirely new laboratory CLIA-certified. Establishing the laboratory and getting certified takes time and costs money. It is not surprising, therefore, that in those SACGHS case studies with an exclusive rights holder (in testing for hemochromatosis, Alzheimer’s disease, BRCA, long-QT, hearing loss, and Canavan’s), the exclusive licensee was beaten to market by multiple university and/or national reference laboratories already offering other genetic tests, which were already CLIA-certified and did not need a patent incentive to add a new test. Indeed, the firm with exclusive rights shut down those competing laboratories already offering a genetic test by threatening enforcement of exclusive patent rights. In this way, the use of patents for diagnostics differs markedly from that for therapeutics and instruments. Universities do not make drugs, vaccines, or laboratory instruments; they do have to offer genetic tests as part of their clinical services that grow out of research, and they can quickly and inexpensively add new tests when new disease associations are published in the literature.
Genetic testing based on DNA sequence technology is thus a highly unusual “market,” because the science that gives rise to new tests is generally conducted at academic centers, and offering new genetic tests is relatively easy for several academic clinical laboratories as well as national reference laboratories that have developed similar tests. The barriers to entry for a single genetic test are relatively low for laboratories already offering other genetic tests, and yet a dedicated startup firm offering just one or a few tests indeed faces significant initial investment hurdle and thus higher marginal costs of production; and university laboratories are “competitors.” One irony is that some of the business models entail academic research institutions exclusively licensing rights for genetic tests to firms that then enforce those rights against laboratory services at other universities.
The relatively low barriers to market entry for new genetic tests could change in the future for several reasons. If the FDA were to regulate genetic tests and require extensive data as part of the process, any laboratory offering a test would have to do clinical studies to produce those data. Payers (including the Centers for Medicare and Medicaid Services, or CMS) could likewise begin to demand evidence of clinical utility of a test before covering it and deciding reimbursement rates, which would again entail up-front expenditures on clinical studies to demonstrate clinical value. If either regulation or payment began to require expensive clinical studies, then the creation of patent incentives to induce R&D investment to overcome market barriers to entry would arise, and the economic model would more closely resemble therapeutics. Patents would not be the only solution to this problem, however, as Congress could instead give the FDA or CMS authority to give exclusive rights to data presented to the agency for approval of a genetic test. If a firm paid for the clinical studies, other firms could not use those data to get a competing product approved (FDA) or paid for (CMS) until after the period of exclusivity. This creates the same kind of incentive as patents but only for that particular use.
Challenges Ahead
Full-genome sequencing
It is just as well that scholars are thinking about creative ways to address potential problems of patent thickets, because the technology for genetic testing is on the verge of being radically altered by the many new technologies for sequencing an entire genome, including a human genome. Complete Genomics published several full-genome sequences in Science in November 2009. The marginal costs of reagents for the least expensive of these full-genome sequences was less than $1,726 (66). This is only a rough indicator of a rapidly evolving technology, and such sequencing is not accurate enough for clinical use, but it does suggest that sometime in the foreseeable future, the cost of deriving an individual’s entire genomic sequence may be less expensive than current prices of genetic testing for conditions such as long-QT syndrome ($5,400 from PGxHealth), BRCA1 and BRCA2 ($3,120 from Myriad), or a spinocerebellar ataxia panel (over $7,000 from Athena). This could change the clinical decision pathway for genetic diagnostics, introducing full-genome sequencing in order to identify possible disease-associated mutations for confirmation in more specific genetic testing. That may depend, however, not just on lower prices and wider use of full-genome sequencing technology but also on how patent claims on existing DNA patents are interpreted and enforced.
If the claims of patents are truly “hard to work around” for 15 of 22 clinical conditions examined by Huys et al. (116), then a full-genome sequence analysis would likely infringe most or all of the 35 patents on their table of patents with at least one blocking claim, as well as many other DNA patent claims that have not been so closely studied. If their estimate that 3% of sequence-based claims block diagnostic use, and if their patents are representative of the more than 15,000 sequence-based claims tracked by Hopkins et al. (112), a full-genome sequence analysis would still infringe several hundred patents. Would patent holders sue the sequencing service? Or would they see the full-genome sequence as a welcome point of entry to more frequent use of their exclusively licensed genetic tests? It is unclear how this will play out.
Many solutions are possible. One is that current court cases and evolving jurisprudence will invalidate the kinds of claims that would be infringed by full-genome sequence analysis. Business practices of patent rights holders could adapt to the new technology, by refraining from literal enforcement or by creating ways to extract small revenue streams from large volumes of full-sequence tests. Patent rights could be incorporated into patent pools or intellectual property collectives analogous to the copyright clearinghouses for songs played over the radio or in commercial establishments, which create revenue streams for rights holders.
ENCODE issues
The changing understanding of how DNA works will confront a legacy of claims in existing DNA patents and the lag in translating new scientific understanding into patent jurisprudence. It turns out that the genome is more dynamic than the collection of “genes” that Johanssen posited in 1913 to explain particulate transmission of inherited characters (121), or that Tatum and Beadle used to explain their “one gene, one enzyme” paradigm in 1941 (27). The conception of a gene has changed subtly but substantially over the past century (123, 142), and patent claims granted on DNA reflect this growth in understanding over time. Yet patent claims can freeze that understanding during the period for which they are granted. A claim to one full-length gene used to produce a therapeutic protein may not be problematic if that patent enables production and expires before other competing products based on component sequences are discovered. The Encyclopedia of DNA Elements (ENCODE) project and other studies are revealing a cybernetic complexity in the control of DNA expression (75). ENCODE’s army of authors concludes its first major publication by observing in understated tones that “the simple view of the genome as having a defined set of isolated loci transcribed independently does not seem to be accurate” (75, p. 812).
Patents are unlikely to stop the exploration of scientific frontiers opening up to genomic technologies. They could, however, do mischief at the margins. The problem of patent scope looms in the background, beyond the possibility of patent thickets and anticommons problems already noted above. Many DNA patents include claims to sequences that “comprise” larger sequences, or to any DNA sequence variation of a claimed sequence regardless of the method used to identify it. Will claims on previously discovered DNA sequences cover those whose functions are just being uncovered, or new purposes not foreseeable when a gene was first discovered? How will the legacy of DNA patents affect the world of micro-RNAs, enhancers, isolators, promoters, silencers, and transcription start sites whose functions are associated with relatively short sequences (but are also dependent on three-dimensional shape, folding, and binding of proteins and RNAs)? To what extent will patent offices allow claims to enclose domains not fully explored at the time a patent is granted?
Patent scope: Ariad v. Lilly
This issue of patent scope is addressed in a recent case, Ariad v. Lilly (19), argued before the full U.S. Court of Appeals for the Federal Circuit in December 2009, which could be appealed to the Supreme Court. The patent in question, US Patent No. 6,410,516 (26), was granted to an extremely distinguished group of fourteen inventors, including Nobel laureates David Baltimore and Phillip Sharp as well as Thomas Maniatis, a Titan of genetics. The patent was assigned to Harvard, MIT, and the Whitehead Institute for Biomedical Research, all vaunted institutions of genetics and genomics. The patent covers “nuclear factors associated with transcriptional regulation,” the NF kappa B pathway, which is involved in control of expression for hundreds of genes. The initial patent application was filed in January 1986 and the patent issued in June 2002. This sixteen-year pendency covers a complex patent prosecution. The patent describes the pathway; at issue now are claims about altering that pathway, exemplified by claim one, “a method for inhibiting expression, in a eukaryotic cell, of a gene whose transcription is regulated by NF-KB, the method comprising reducing NF-KB activity in the cell such that expression of said gene is inhibited.” Every one of the 203 claims granted in this patent begins with “method of” or “method for” language, so these are not structural claims but claims intended to cover any means of achieving the specified functions. At stake was whether having discovered the pathway, these research institutions should be able to tap revenues from the sale of drugs that affect the NF kappa B pathway.
The crucial interpretation in this case was of Section 112 of the U.S. Patent Act, the “written description” requirement (227). At least twenty-five amicus briefs were submitted to the court. Genome scientists will find the brief from the Universities of California and Texas, the Wisconsin Alumni Research Foundation (WARF), and several other universities of particular interest (171). Between the litigants and the list of research institutions in this brief, most of the top academic DNA patent holders were involved in this case. WARF noted it had earned over $1 billion from licensing fees to fund research. Its brief centered on the value of patent rights to “pioneering biological inventions” arising in university research. It argued for “recruitment of commercial entities willing to undertake the huge investments necessary to refine and develop foundational university research into medical and biopharmaceutical products,” although in this case Lilly made just such investments independent of the patents, well before the patent issued, and it is quite clear that patents were not needed to pursue applications of the NF kappa B pathway. Indeed, drug function was not precisely known at the time the compounds at stake in this case were first explored as therapeutics. The university amicus brief asserts, demonstrably falsely in this case, that denial of patent protection would keep “important and possibly life-saving advancements out of the public’s reach” but, more to the point, would deprive “universities and research institutions of the opportunity to generate funds for continued scientific research, education, and innovation” (171, p. 9). The deep irony of this position is clear and addressed in the 2005 National Academies report that surveyed scientists who study the NF kappa B pathway (219). The research that academic scientists studying NF kappa B conducted clearly infringed one or more of the 203 claims in US Patent No. 6,410,516. In arguing for broad claims on “pioneering biological inventions” to enable collection of royalty streams, the brief thus implies but does not state explicitly that issuing such claims can avoid harm to research only with discretionary and highly selective enforcement of the resulting exclusive rights—most notably, not enforcing patents against university researchers, but by implication commercial users are fair game. The research system works because university scientists pervasively ignore the rights that technology licensing officials at the same universities argue are necessary to generate income.
A story that began with university research of fundamental scientific value, as well as considerable commercial promise, has led to patents on seminal recombinant DNA technology in 1980 and now to a pending appellate court decision about the breadth of patent rights to a pleiotrophic transcriptional pathway. Universities find themselves speaking out of both sides of their mouths, arguing for the virtues of commercialization and the social value of exclusive rights that their own scientists routinely ignore. The resolution of this hypocrisy can come only from more explicit policies that address the nuances of patenting and licensing DNA inventions, and that accommodate the rights and interests of universities as users of patented inventions as well as holders of patent rights. The best hope is for a process in which the scientific and academic functions of academic research institutions find common ground with their business interests.
Figure 5.
Instances and outcomes of human gene patent litigation. Christopher Holman searched Lexis-Nexis (http://www.lexis.com) databases and federal court cases for litigated patents that included either the term SEQ ID NO. in the claims or terms used in the DNA Patent Database query in the claims or abstract. He also searched the Westlaw Intellectual Property Docket (http://www.westlaw.com) for litigation involving patents in Jensen & Murray’s patents database (J&M Patents) that included terms from the DNA Patent Database in the abstract. The number of patents identified in J&M Patents is indicated in parentheses (120). Data through April 2007 is from Reference (109) and used with the authors’ permission. Holman’s data do not include the pending case concerning BRCA sequences and testing methods (20, 21A).
ACKNOWLEDGEMENTS
The authors wish to thank several scholars who generously gave permission to use their data in this article, including Tim Caulfield, Tania Bubela and C. J. Murdoch of the University of Alberta, Michael Hopkins of the University of Sussex, Christopher Holman of the Law School for the University of Missouri, Kansas City, and LeRoy Walters and Mara Snyder of the DNA Patent Database, Georgetown University. Richard Gold and Julia Carbone were fully part of our research team, indeed our extended family, even before they became a family through matrimony. Our thanks go to the many students and colleagues, particularly those who contributed to genetic testing case studies (Subhashini Chandrasekharan, Misha Angrist, Alessa Colaianni, Katie Skeehan, Ashton Powell, Tamara James, Emily Pitlick, Christopher DeRienzo, Melissa Fiffer, and Christopher Conover). Geertrui van Overwalle and her colleagues at the University of Leuven contributed insights into diagnostic uses of gene patents from across the Atlantic. Daniel Kevles, Sally Smith Hughes, and Nicholas Rasmussen provided unpublished material. James Evans of the University of North Carolina and Rochelle Dreyfus of New York University worked tirelessly for the task force on the effect of patenting and licensing on clincal access to genetic testing for the Secretary’s Advisory Committee on Genetics, Health and Society, with its able staff that included Sarah Carr, Yvette Seger and Darren Greninger. Rebecca Eisenberg, Arti Rai, James Boyle, Jerome Reichman, Bhaven Sampat, Anthony So and many others too numerous to mention have provided many ideas and critiques and are dearly appreciated as stalwart colleagues. This article draws heavily on research funded by the National Human Genome Research Institute and US Department of Energy through a grant, P50 HG003391.
Footnotes
The other genomic start-up firms were Darwin Molecular, Mercator, Myriad, Millennium, Sequana, and Genome Therapeutics, which changed its name from Collaborative Genetics, indicating a shift from finding and mapping genetic linkage markers toward sequencing and other genomic technologies (52).
Funders included the National Human Genome Research Institute, the Japanese Ministry of Education, Culture, Sports, Science, and Technology, the Wellcome Trust, Nuffield Trust, Wolfson Foundation, the United Kingdom’s EPSRC, Genome Canada, Genome Quebec, the Chinese Academy of Sciences, the Ministry of Science and Technology of the People’s Republic of China, the National Natural Science Foundation of China, the Hong Kong Innovation and Technology Commission, the University Grants Committee of Hong Kong, the SNP Consortium, the W.M. Keck Foundation, and the Delores Dore Eccles Foundation (119).
DISCLOSURE STATEMENT
One author (RC-D) served on the National Research Council study of University Management of Intellectual Property (2008–2010) and is a faculty advisor to the Duke (and occasionally University of North Carolina) chapters of Universities Allied for Essential Medicines. Both authors did commissioned research for the Secretary’s Advisory Committee for Genetics, Health and Society 2007–2010. The Center for Genome Ethics, Law and Policy where both authors work has accepted no corporate funding for research since its creation in 2002. Funding is provided by federal grants, grants from nonprofit philanthropies and university sources, and an intial grant from The Duke Endowment.
LITERATURE CITED
- 1.Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, et al. Complementary DNA sequencing: expressed sequence tags and Human Genome Project. Science. 1991;252(5013):1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 2.Adelman D. A fallacy of the commons in biotech patent policy. Berkeley Tech. L. J. 2005;20(2):985–1030. [Google Scholar]
- 3.Adelman D. The irrationality of speculative gene patents. In: Libecap G, editor. Advances in the Study of Entrepreneurship, Innovation and Economic Growth. Vol. 16: University Entrepreneurship and Technology Transfer: Process, Design and Intellectual Property. Oxford, UK: Elsevier; 2005. pp. 123–154. [Google Scholar]
- 4.Adelman D, DeAngelis K. Patent metrics: the mismeasure of innovation in the biotech patent debate. Tex. L. Rev. 2007;85(7):1677–1744. [Google Scholar]
- 5.Adler R. Genome research: fulfilling the public’s expectations for knowledge and commercialization. Science. 1992;257(5072):908–914. doi: 10.1126/science.1502557. [DOI] [PubMed] [Google Scholar]
- 6.Affymetrix infringed Ed Southern’s microarray patent. Biotech Patent News. 2000 Nov. 1. http://www.entrepreneur.com/tradejournals/article/67939117.html.
- 7.Albainy-Jenei S. BulletProof: interview with ACLU attorney Chris Hansen over gene patents. 2009 http://www.patentbaristas.com/archives/2009/11/12/bulletproof-interview-with-aclu-attorney-chris-hansen-over-gene-patents.
- 8.Alberts B. Dangers in EST patent law. 1997. Plant Mol. Biol. Rep. 1997;15(3):205–208. [Google Scholar]
- 9.Altman LK. A new insulin given approval for use in US. New York Times. 1982 Oct 30;:A1. [Google Scholar]
- 9A.Amgen Inc. Appeals court affirms patent infringement ruling against Roche’s peg-EPO product. 2009 Press release. http://www.amgen.com/media/media_pr_detail.jsp?year=2009&releaseID=1332195.
- 10.Amgen Inc. v. Chugai Pharmaceutical Co. Ltd. et al. 13 U.S.P.Q.2d 1737 (D. Mass 1989) [Google Scholar]
- 11.Amgen Inc. v. Chugai Pharmaceutical Corporation and Genetics Institute. 927 F.2d 1200 (Fed. Cir 1991) [Google Scholar]
- 12.Amgen Inc. v. F. Hoffmann-La Roche Ltd. et al. No. 05-12237-WGY (D. Mass. Oct. 2, 2008) [Google Scholar]
- 13.Amgen Inc. v. F. Hoffmann-La Roche Ltd. et al. Nos. 2009-1020 and 1020–1096 (Fed. Cir. Sept. 15, 2009) [Google Scholar]
- 14.Angrist M, Chandrasekharan S, Heaney C, Cook-Deegan R. Impact of gene patents and licensing practices on access to genetic testing for Long QT syndrome. Genet. Med. 2010;12(4):S111–S154. doi: 10.1097/GIM.0b013e3181d68293. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ankeny R. The natural history of Caenorhabditis elegans research. Nat. Rev. Genet. 2001;2(6):474–479. doi: 10.1038/35076538. [DOI] [PubMed] [Google Scholar]
- 16.Ankeny R. Sequencing the genome from nematode to human: changing methods, changing science. Endeavor. 2003;27(2):87–92. doi: 10.1016/s0160-9327(03)00061-9. [DOI] [PubMed] [Google Scholar]
- 17.Amgen, Inc. Appeals court affirms patent infringement ruling against Roche’s peg-EPO product. 2009 http://www.amgen.com/media/media_pr_detail.jsp?year=2009&releaseID=1332195.
- 18.Applera Corporation and Roche Molecular Systems, Inc. v. MJ Research Inc. and Michael and John Finney. No. 3:98cv1201 (JBA)(D. Conn. 2005) [Google Scholar]
- 19.Ariad Pharmaceuticals, Inc., et al. v. Eli Lilly and Co. No. 2008--1248 (Fed. Cir. Dec. 7, 2009) [Google Scholar]
- 20.Association for Molecular Pathology, et al. v. United States Patent and Trademark Office et al. Civil Action No. 09--4515 (RWS)(S.D.N.Y., filed May 12, 2009) [Google Scholar]
- 21.Association for Molecular Pathology et al. v. United States Patent and Trademark Office et al. Civil Action No. 09--4515 (RWS)(S.D.N.Y. Nov. 1, 2009) (order denying defendants’ motion to dismiss) [Google Scholar]
- 21A.Association for Molecular Pathology, et al., v. United States Patent and Trademark Office, et al. No. 09—CV--4515 (RWS) (S.D.N.Y. March 29, 2010) (opinion granting plaintiffs’ motion for summary judgment in part, denying Myriad Genetics’ motion for summary judgment, granting United States Patent and Trademark Office’s motion for judgment on the pleadings, and declaring claims-in-suit invalid) [Google Scholar]
- 22.Australian Law Reform Commission. Report 99---Genes and Ingenuity: Gene Patenting and Human Health. Sydney: Australian Law Reform Commission; 2004. http://www.austlii.edu.au/au/other/alrc/publications/reports/99/ [Google Scholar]
- 23.Au-Young J, Bandman O, Hawkins PR, Wilde CG. No. 5,817,479. US Patent. 1996
- 24.Ayme S, Matthijs G, Soini S on behalf of the ESHG Working Party on Patenting and Licensing. Patenting and licensing in genetic testing: recommendations of the European Society of Human Genetics. Eur. J. Hum. Genet. 2008;16(4):405–411. doi: 10.1038/sj.ejhg.5201929. [DOI] [PubMed] [Google Scholar]
- 25.Ayukawa Y, Shinya S, Tamura M. No. 3615654. US Patent. 1971
- 26.Baltimore D, Sen R, Sharp PA, Singh H, Staudt L, et al. No. 6,410,516. US Patent. 2002
- 27.Beadle G, Tatum E. Genetic control of biochemical reactions in neurospora. Proc. Natl. Acad. Sci. USA. 1941;27(11):499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berkowitz A, Kevles D. Patenting human genes: the advent of ethics in the political economy of patent law. In: Magnus D, Caplan A, McGee G, editors. Who Owns Life? Amherst, NY: Prometheus Books; 2007. pp. 75–97. [Google Scholar]
- 29.Berman EP. Why did universities start patenting? Institution-building and the road to the Bayh-Dole Act. Soc. Stud. Sci. 2008;38(6):835–871. doi: 10.1177/0306312708098605. [DOI] [PubMed] [Google Scholar]
- 30.Bilski v. Kappos. 556 U.S. ___(2009) [Google Scholar]
- 31.Bok D. Universities in the Marketplace: The Commercialization of Higher Education. Princeton, NJ: Princeton Univ. Press; 2004. [Google Scholar]
- 32.Brody B. Intellectual property and biotechnology: the U.S. internal experience---part I. Kennedy Inst. Ethics J. 2006;16(1):1–38. doi: 10.1353/ken.2006.0002. [DOI] [PubMed] [Google Scholar]
- 33.Brody B. Intellectual property and biotechnology: the U.S. internal experience---part II. Kennedy Inst. Ethics J. 2006;16(2):105–128. doi: 10.1353/ken.2006.0007. [DOI] [PubMed] [Google Scholar]
- 34.Brody B. Intellectual property and biotechnology: the European debate. Kennedy Inst. Ethics J. 2007;17(2):69–110. doi: 10.1353/ken.2007.0008. [DOI] [PubMed] [Google Scholar]
- 35.Buchanan JM, Yoon YJ. Symmetric tragedies: commons and anticommons. J. Law Econ. 2000;43(1):1–13. [Google Scholar]
- 35B.Bugbee B. Genesis of American Patent and Copyright. Washington, DC: Public Affairs Press; 1964. [Google Scholar]
- 36.Bunn H. End run around EPO. N. Engl. J. Med. 2009;361(19):1901–1903. doi: 10.1056/NEJMe0908186. [DOI] [PubMed] [Google Scholar]
- 36A.Cancer Voices Australia. (ABN 93 322 703 427) & Anor v Myriad Genetics Inc & Ors (Federal Court of Australia, New South Wales Registry, filed June 8, 2010) [Google Scholar]
- 37.Caskey T, Tribble J. Who owns the code? Science and Public Affairs. 1997:52–55. Autumn. [Google Scholar]
- 38.Caulfield T, Bubela T, Murdoch C. Myriad and the mass media: the covering of a gene patent controversy. Genet. Med. 2007;9(12):850–855. doi: 10.1097/gim.0b013e31815bf965. [DOI] [PubMed] [Google Scholar]
- 39.Caulfield T, Cook-Deegan R, Kieff F, Walsh J. Evidence and anecdotes: an analysis of human gene patenting controversies. Nat. Biotechnol. 2005;24(9):1091–1094. doi: 10.1038/nbt0906-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandrasekharan S, Fiffer M. Impact of gene patents and licensing practices on access to genetic testing for hearing loss. Genet. Med. 2010;12(4):S155–S170. doi: 10.1097/GIM.0b013e3181d7b053. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandrasekharan S, Heaney C, James T, Conover C, Cook-Deegan R. Impact of gene patents and licensing practices on access to genetic testing for cystic fibrosis. Genet. Med. 2010;12(4):S194–S211. doi: 10.1097/GIM.0b013e3181d7cf7d. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandrasekharan S, Pitlick E, Heaney C, Cook-Deegan R. Impact of gene patents and licensing practices on access to genetic testing for hereditary hemochromatosis. Genet. Med. 2010;12(4):S156–S171. doi: 10.1097/GIM.0b013e3181d7acb0. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho M, Illangasekare S, Weaver M, Leonard D, Merz J. Effects of patents and licenses on provision of clinical genetic testing services. J. Mol. Diagn. 2003;5(1):3–8. doi: 10.1016/S1525-1578(10)60444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cockburn I, Henderson R. Public-private interaction in pharmaceutical research. Proc. Natl. Acad. Sci. USA. 1996;93(23):12725–12730. doi: 10.1073/pnas.93.23.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen SN, Boyer HW. No. 4,237,224. US Patent. 1980
- 46.Cohen SN, Chang AC, Boyer HW, Helling R. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA. 1973;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen WM, Nelson RR, Walsh JP. Links and impacts: the influence of public research on industrial R&D. Manag. Sci. 2002;48(1):1–23. [Google Scholar]
- 48.Colaianni A, Chandrasekharan S, Cook-Deegan R. Impact of gene patents and licensing practices on access to genetic testing for Tay-Sachs and Canavan disease. Genet. Med. 2010;12(4):S5–S14. doi: 10.1097/GIM.0b013e3181d5a669. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colaianni A, Cook-Deegan R. Columbia University’s Axel patents: technology transfer and implications for the Bayh--Dole Act. Milbank Q. 2009;87(3):683–715. doi: 10.1111/j.1468-0009.2009.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colyvas J, Crow M, Gelijns A, Mazzoleni R, Nelson RR. How do university inventions get into practice? Manag. Sci. 2002;48(1):61–72. [Google Scholar]
- 51.New York Times, ed. Company News: Affymetrix settles patent disputes with Oxford. New York Times. 2004 March 27;:C4. [Google Scholar]
- 52.Cook-Deegan R. Survey of genome sciences corporations. Washington, DC: Commissioned report, Office of Technology Assessment, US Congress; 1994. http://www.genome.duke.edu/centers/cpg/archives/historical/ [Google Scholar]
- 53.Cook-Deegan R. The Gene Wars: Science, Politics and the Human Genome. New York: WW Norton; 1994. http://www.genome.duke.edu/books/gene-wars/ [Google Scholar]
- 54.Cook-Deegan R. Sequence upon sequence. 1994:56–77. See ref. 53. [Google Scholar]
- 55.Cook-Deegan RM, DeRienzo C, Carbone J, Chandrasekharan S, Heaney C, Conover C. Impact of gene patents on access to genetic testing for inherited susceptibility to cancer: comparing breast and ovarian cancers to colon cancers. Genet. Med. 2010;12(4):S15–S38. doi: 10.1097/GIM.0b013e3181d5a67b. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crichton M. Next. New York: Harper Collins; 2006. [Google Scholar]
- 57.Crichton M. Patenting life. New York Times. 2007 February 13;:A23. [Google Scholar]
- 58.Danish Council of Ethics. Patenting human genes and stem cells. Copenhagen: Danish Council of Ethics; 2004. http://etiskraad.synkron.com/graphics/03_udgivelser/engelske_publikationer/patenting_human_genes/patents04/index.htm. [Google Scholar]
- 59.Delphion Patent Database. 2009 http://www.delphion.com.
- 60.Diamond v. Chakrabarty. 447 U.S. 303. 1980 [PubMed] [Google Scholar]
- 61.Dickinson Q. Revised utility examination guidelines; request for comments. Fed. Regist. 1999;64:71440–71442. [Google Scholar]
- 62.Dickinson Q. Guidelines for examination of patent applications under the 35 U.S.C. 112, ¶ 1, “Written Description” requirement. Fed. Regist. 2001;66:1099–1111. [Google Scholar]
- 63.Dickinson Q. Utility examination guidelines. Fed. Regist. 2001;66:1092–1099. [Google Scholar]
- 64.DNA Patent Database. http://dnapatents.georgetown.edu (Algorithm used to located DNA patents available at http://dnapatents.georgetown.edu/SearchAlgorithm-Delphion-20030512.htm)
- 65.Doll J. The patenting of DNA. Science. 1998;280(5364):689–690. doi: 10.1126/science.280.5364.689. [DOI] [PubMed] [Google Scholar]
- 66.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 67.eBay, Inc. v. MercExchange, LLC. 547 U.S. 388. 2006. [Google Scholar]
- 68.Eisenberg RS. Intellectual property at the public-private divide: the case of large-scale cDNA sequencing. U. Chi. L. Sch. Roundtable. 1996;3(2):557–573. [Google Scholar]
- 69.Eisenberg RS. Intellectual property issues in genomics. Trends Biotechnol. 1996;14(8):203–307. doi: 10.1016/0167-7799(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 70.Eisenberg RS. Public research and private development: patents and technology transfer in government-sponsored research. Va. Law Rev. 1996;82(8):1663–1727. [Google Scholar]
- 71.Eisenberg RS. Noncompliance, nonenforcement, nonproblem? Rethinking the anticommons in biomedical research. Houston L. Rev. 2008;45(4):1059–1099. [Google Scholar]
- 72.Eisenberg RS, Merges RP. Opinion letter as to the patentability of certain inventions associated with the identification of partial DNA sequences. AIPLA Q. J. 1995;23(1):1–52. [Google Scholar]
- 73.Eisenberg RS, Nelson RR. Public versus proprietary science: a fruitful tension? Acad. Med. 2002;77(12):1329–1399. doi: 10.1097/00001888-200212001-00011. [DOI] [PubMed] [Google Scholar]
- 74.Eisenberg RS, Rai A. Harnessing and sharing the benefits of state-sponsored research: intellectual property rights and data sharing in California’s stem cell initiative. Berkeley Tech. L. J. 2006;21(3):1187–1213. [Google Scholar]
- 75.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.European Union. Directive 98/44/EC of the European Parliament on the Legal Protection of Biotechnological Inventions. Off. J. Eur. Communities. 1998 L.213 30/07/1998 0013. [Google Scholar]
- 77.Ex parte Kubin. 83 U.S.P.Q.2d 1410 (Bd. Pat. App. & Int 2007) [Google Scholar]
- 78.Feldman M, Colaianni A, Liu CK. Lessons from the commercialization of the Cohen--Boyer patents: the Stanford University licensing program. In: Krattiger A, Mahoney RT, Nelsen L, Thomson JA, Bennett AB, et al., editors. Intellectual Property Management in Health and Agricultural Innovation: A Handbook of Best Practices. Oxford, UK: MIHR; 2007. pp. 1797–1808. Davis, CA: PIPRA. http://www.ipHandbook.org. [Google Scholar]
- 79.Fore J, Wiechers I, Cook-Deegan R. The effects of business practices, licensing, and intellectual property on development and dissemination of the polymerase chain reaction: case study. J. Biomed. Discov. Collab. 2006;1(7):1–17. doi: 10.1186/1747-5333-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Form 10-K Annual Report. 2007 http://www.sec.gov/Archives/edgar/data/1110803/000093639207000158/a27675e10vk.htm.
- 81.Form 10-K Annual Report. 2008 http://www.sec.gov/Archives/edgar/vprr/09/9999999997-09-016377.
- 82.Form 10-K Annual Report. 2008 http://www.sec.gov/Archives/edgar/data/1110803/000093639208000144/a38423e10vk.htm.
- 83.Form 10-K Annual Report. 2009 http://www.sec.gov/Archives/edgar/data/1110803/000093639209000092/a51300e10vk.htm.
- 84.Fredrickson DS. Letter from Donald S. Fredrickson to Dr. Robert M. Rosenzweig. Recomb. DNR Res. 1977;2:1A–1B. [Google Scholar]
- 85.Fredrickson DS. Sample letter on DHEW patent policy as applied to recombinant DNA inventions. Recomb. DNA Res. 1977;2:21–24. [Google Scholar]
- 86.Fredrickson DS. The Recombinant DNA Controversy: A Memoir, Science, Politics, and the Public Interest 1974--1981. Washington, DC: ASM Press; 2001. [Google Scholar]
- 87.Gelfand DH, Stoffel S, Lawyer FC, Saiki RK. No. 4,889,818. US Patent. 1989
- 88.Gerhold D, Caskey C. It’s the genes! EST access to human genome content. BioEssays. 1996;18(12):973–981. doi: 10.1002/bies.950181207. [DOI] [PubMed] [Google Scholar]
- 89.Germann C. The Swiss approach to compulsory licensing for diagnostic products and processes. In: van Overwalle G, editor. Gene Patents and Public Health. Brussels: Bruylant; 2007. pp. 149–157. [Google Scholar]
- 90.Gershon D. Amgen scores a knockout. Nature. 1991;350(6314):99. doi: 10.1038/350099a0. [DOI] [PubMed] [Google Scholar]
- 91.Gilbert W. Interview with M Nicholson, R Cook-Deegan. Durham, NC: Center for Public Genomics, Duke University; 2005. http://dukespace.lib.duke.edu/dspace/handle/10161/1561. [Google Scholar]
- 92.Gilham PT, Weith HL. No. 3,730,844. US Patent. 1973
- 93.Gitschier J. Wonderful life: an interview with Herb Boyer. PLoS Genet. 2009;5(9) doi: 10.1371/journal.pgen.1000653. e1000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gold ER, Carbone J. Myriad Genetics: in the eye of the policy storm. Genet. Med. 2010;12(4):S39–S70. doi: 10.1097/GIM.0b013e3181d72661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gold ER, Gallochat A. The European Biotech Directive: past as prologue. Eur. J. Law. 2001;7(3):331–366. [Google Scholar]
- 96.Goldstein J. Critical analysis of patent pools. In: van Overwalle G, editor. Gene Patents and Collaborative Licensing Models: Patent Pools, Clearinghouses, Open Source Models and Liability Regimes. Cambridge, UK: Cambridge Univ. Press; 2009. pp. 57–59. [Google Scholar]
- 97.Greenberg DS. Science for Sale: The Perils, Rewards and Delusions of Campus Capitalism. Chicago: Univ. of Chicago Press; 2007. [Google Scholar]
- 98.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 99.Hall S. Invisible Frontiers: The Race to Synthesize the Human Gene. Boston: Atlantic Monthly Press; 1987. [Google Scholar]
- 100.Hardee ME, Arcasoy MO, Blackwell KL, Kirkpatrick JP, Dewhirst MW. Erythropoietin biology in cancer. Clin. Cancer Res. 2006;12(2):332–339. doi: 10.1158/1078-0432.CCR-05-1771. [DOI] [PubMed] [Google Scholar]
- 101.Healy B. On gene patenting. N. Engl. J. Med. 1992;327(9):664–668. doi: 10.1056/NEJM199208273270930. [DOI] [PubMed] [Google Scholar]
- 102.Heller M. The tragedy of the anticommons: property in the transition from Marx to markets. Harvard Law Rev. 1998;111(3):621–688. [Google Scholar]
- 103.Heller M, Eisenberg R. Can patents deter innovation? The anticommons in biomedical research. Science. 1998;280(5364):698–701. doi: 10.1126/science.280.5364.698. [DOI] [PubMed] [Google Scholar]
- 104.Henry Huang v. California Institute of Techology et al. Case CV 03--1140 (C.D. Cal. 2004) [Google Scholar]
- 105.Hoffman-La Roche, Inc. et al. v. Promega Corp. 1994 U.S. Dist. LEXIS 10174 (N.D. Cal. 1994) [Google Scholar]
- 106.Hoffmann-La Roche, Inc. v. Promega Corp. 323 F.3d 1354 (Fed. Cir. 2003) [Google Scholar]
- 107.Hoffmann-La Roche, Inc. et al. v. Promega Corp. 319 F.Supp.2d 1011 (N.D. Cal. 2004) [Google Scholar]
- 108.Holman CM. The impact of human gene patents on innovation and access: a survey of human gene patent litigation. UMKC Law Rev. 2007;76(2):295–361. [Google Scholar]
- 109.Holman CM. Trends in human gene patent litigation. Science. 2008;322(5899):198–199. doi: 10.1126/science.1160687. [DOI] [PubMed] [Google Scholar]
- 110.Hood L, Hunkapiller M, Smith L. Automated DNA sequencing and analysis of the human genome. Genomics. 1987;1(3):201–212. doi: 10.1016/0888-7543(87)90046-2. [DOI] [PubMed] [Google Scholar]
- 111.Hopkins M, Mahdi S, Patel P, Thomas S. DNA patenting: the end of an era? Nat. Biotechnol. 2007;25(2):185–187. doi: 10.1038/nbt0207-185. [DOI] [PubMed] [Google Scholar]
- 112.Hopkins M, Mahdi S, Thomas S, Patel P. The Patenting of Human DNA: Global Trends in Public and Private Sector Activity (the PATGEN project) Falmer, Brighton, East Sussex, United Kingdom: Science and Technology Policy Research; 2006. [Google Scholar]
- 113.Huang KG, Murray FE. Does patent strategy shape the long-run supply of public knowledge? Evidence from human genetics. Acad. Management J. 2009;52(6):1193–1221. [Google Scholar]
- 114.Hughes SS. Making dollars out of DNA: the first major patent in biotechnology and the commercialization of molecular biology 1974--1980. Isis. 2001;92(3):541–575. doi: 10.1086/385281. http://digitalassets.lib.berkeley.edu/roho/ucb/text/makingdollarsoutofdna.pdf. [DOI] [PubMed]
- 115.Hughes SS. Genes, Genentech and the Rise of Commercial Biotechnology. Chicago: Chicago Univ. Press; 2010. In Press. [Google Scholar]
- 116.Huys I, Berthels N, Matthijs G, van Overwalle G. Legal uncertainty in the areas of genetic diagnostic testing. Nat. Biotechnol. 2009;27(10):903–909. doi: 10.1038/nbt1009-903. [DOI] [PubMed] [Google Scholar]
- 117.Illumina countersues Life Technologies for patent infringement. 2009 http://investor.illumina.com/phoenix.zhtml?c=121127&p=irol-newsArticle&ID=1342003&highlight=
- 118.In re Deuel. 51 F.3d 1552 (Fed. Cir. 1995) [Google Scholar]
- 119.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jensen K, Murray F. Intellectual property landscape of the human genome. Science. 2005;310(5746):239–240. doi: 10.1126/science.1120014. [DOI] [PubMed] [Google Scholar]
- 121.Johannsen W. Elemente der Exakten Erblichkeitslehre mit Grundzügen der Biologischen Variationsstatistik. Jena, Germany: Gustav Fisher; 1913. [Google Scholar]
- 122.Junod SW. Celebrating a milestone: FDA’s approval of first genetically-engineered product. Update. 2007 Sept.--Oct;:43–44. [Google Scholar]
- 123.Keller E. The Century of the Gene. Cambridge, MA: Harvard Univ. Press; 2000. [Google Scholar]
- 124.Kevles DJ. Ananada Chakrabarty wins a patent: biotechnology, law and society, 1972--1980. Hist. Stud. Phys. Biol. 1994;25(1):111–135. [PubMed] [Google Scholar]
- 125.Kiley T. Patents on random complementary DNA fragments? Sciences. 1992;257(5072):915–918. doi: 10.1126/science.1502558. [DOI] [PubMed] [Google Scholar]
- 126.Kirin-Amgen Inc. and others (Appellants) v. Hoechst Marion Roussel Limited and others (Respondents); Kirin-Amgen Inc and others (Respondents) v. Hoechst Marion Roussel Limited and others (Appellants) (Conjoined Appeals) UKHL 46 on appeal from: [2002] EWCA Civ 1096. 2004. [Google Scholar]
- 127.Krimsky S. Genetic Alchemy: The Social History of the Recombinant DNA Controversy. Cambridge, MA: MIT Press; 1984. [Google Scholar]
- 128.Krimsky S. Science in the Private Interest: Has the Lure of Profits Corrupted Biomedical Research? Lanham, MD: Rowman and Littlefield; 2003. [Google Scholar]
- 129.KSR International Co. v. Teleflex Inc. 550 U.S. 398. 2007. [Google Scholar]
- 130.Lab Corp v. Metabolite Laboratories, Inc. 548 U.S. 124. 2006. [Google Scholar]
- 131.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 132.Lenoir T, Giannella E. The emergence and diffusion of DNA microarray technology. J. Biomed. Discov. Collab. 2006;1(11):1–39. doi: 10.1186/1747-5333-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Life Technologies initiates suit against Illumina alleging patent infringement. 2009 http://investor.illumina.com/phoenix.zhtml?c=121127&p=irol-newsArticle&ID=1335039&highlight=
- 134.Litigation settled on gene patents. New York Times. 2001 Dec 22;:C14. [Google Scholar]
- 135.Macilwain C. Gene patent study drowned as OTA sinks. Nature. 1995;376(6541):541. doi: 10.1038/376541a0. [DOI] [PubMed] [Google Scholar]
- 136.Marshall E. A bitter battle over insulin gene. Science. 1997;227(5329):1028–1030. doi: 10.1126/science.277.5329.1028. [DOI] [PubMed] [Google Scholar]
- 137.Marshall E. Courts take a narrow view of UC’s claims. Science. 1997;227(5329):1029. [Google Scholar]
- 138.Maskus K. Intellectual Property Rights in the Global Economy. Washington, DC: Institute for International Economics; 2000. pp. 15–26. [Google Scholar]
- 139.Memorandum of Understanding between Myriad Genetics and the U.S. National Cancer Institute. 1999. Quoted in Ref. 55, note 3. [Google Scholar]
- 140.Merz JF, Henry MR. The prevalence of patent interferences in gene technology. Nat. Biotechnol. 2004;22(2):153–154. doi: 10.1038/nbt0204-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miller C. Letter to Paul Adam Marks. Columbia University: Vice President for Health Services; 1981. http://www.genome.duke.edu/centers/cpg/documents/project1/wigler-silverstein-axel/Columbia%20letter%202-24-1981.pdf. [Google Scholar]
- 142.Morange M. The Misunderstood Gene. Cambridge, MA: Harvard Univ. Press; 2002. [Google Scholar]
- 143.Mossoff A. Rethinking the development of patents: an intellectual history 1550–1800. Hastings L. J. 2001;52(6):1255–1322. [Google Scholar]
- 144.Mossoff A. A Stitch in Time: The Rise and Fall of the Sewing Machine Patent Thicket. Arlington, Virg: Law and Economics Center, George Mason School of Law; 2009. Research Paper No. 09--19. http://ssrn.com/abstract=1354849. [Google Scholar]
- 145.Mowery D, Nelson R, Sampat B, Ziedonis A. The effects of the Bayh--Dole Act on U.S. university research and technology transfer. In: L Branscomb L, editor. Industrializing Knowledge. Cambridge, MA: MIT Press; 1999. pp. 269–306. [Google Scholar]
- 146.Mowery D, Nelson R, Sampat B, Ziedonis A. Ivory Tower and Industrial Innovation: University-Industrial Technology Transfer before and after the Bayh--Dole Act. Stanford, CA: Stanford Business Press; 2004. [Google Scholar]
- 147.Mullin J. Lawsuit looks to “take down” patents on human genes. IP Law & Business. 2009 May 15; http://www.law.com/jsp/law/LawArticleFriendly.jsp?id=1202430726483.
- 148.Mullis KB. No. 4,683,202. US Patent. 1987
- 149.Mullis KB. The unusual origin of the polymerase chain reaction. Sci. Am. 1990;262(4):56–65. doi: 10.1038/scientificamerican0490-56. [DOI] [PubMed] [Google Scholar]
- 150.Mullis KB. PCR and scientific invention: the trial of DuPont v. Cetus. In: Mullis KB, Ferre F, Gibbs RA, editors. PCR: The Polymerase Chain Reaction. Boston: Birkhäuser; 1994. pp. 427–441. [Google Scholar]
- 151.Mullis KB, Erlich HA, Arnheim N, Horn GT, Saiki RK, Scharf SJ. No. 4,683,195. US Patent. 1987
- 152.Murray F, Stern S. Do formal intellectual property rights hinder the free flow of scientific knowledge? An empirical test of the anticommons hypothesis. J. Econ. Behav. Organ. 2007;63(4):648–687. [Google Scholar]
- 153.Myriad Genetics, Inc. v. Oncordmed, Inc. No. 97--922 (C.D. Utah filed Dec. 2, 1997) [Google Scholar]
- 154.Myriad Genetics, Inc. v. Univ. of Pa. No. 98--829 (D.C. Utah filed Nov. 19, 1998) [Google Scholar]
- 155.Nard CA, Morriss AP. Constitutionalizing patents: from Venice to Philadelphia. Rev. L. & Econ. 2006;2(2):223–321. [Google Scholar]
- 156.National Institutes of Health. Recomb. DNR Res. 1977;2:62–155. [Google Scholar]
- 157.National Research Council. Reaping the Benefits of Genomic and Proteomic Research: Intellectual Property Rights, Innovation and Public Health. Washington, DC: National Research Council; 2006. http://www.nap.edu/catalog.php?record_id=11487. [PubMed] [Google Scholar]
- 158.Nelson RR. The market economy and the scientific commons. Res. Policy. 2004;33(3):455–471. [Google Scholar]
- 159.Nuffield Council on Bioethics. The Ethics of Patenting DNA: A Discussion Paper. London: Nuffield Council on Bioethics; 2002. http://www.nuffieldbioethics.org/fileLibrary/pdf/theethicsofpatentingdna.pdf. [Google Scholar]
- 160.Office of Technology Assessment. Impacts of Applied Genetics. Washington, DC: Office of Technology Assessment, US Congress; 1981. http://www.fas.org/ota/reports/8115.pdf. [Google Scholar]
- 161.Office of Technology Assessment. Commercial Biotechnology: An International Analysis. Washington, DC: Office of Technology Assessment, US Congress; 1984. http://www.princeton.edu/~ota/disk3/1984/8407_n.html. [Google Scholar]
- 162.Office of Technology Assessment. Patenting Life. Washington, DC: Office of Technology Assessment, US Congress; 1989. http://www.fas.org/ota/reports/8924.pdf. [Google Scholar]
- 163.Office of Technology Assessment. Biotechnology in a Global Economy. Washington, DC: Office of Technology Assessment, US Congress; 1991. http://www.fas.org/ota/reports/9110.pdf. [Google Scholar]
- 164.Office of Technology Assessment. The Human Genome Project and Patenting DNA Sequences. Washington, DC: Approved prepublication draft. Office of Technology Assessment, US Congress; 1994. http://kie.georgetown.edu/nrcbl/documents/dnapatents/OTAdraft.pdf. [Google Scholar]
- 165.Oncormed, Inc. v. Myriad, Inc. No. 97–2722 (D.D.C. filed Nov. 17, 1997) [Google Scholar]
- 166.Ontario Ministry of Health. Toronto, Canada: Ontario Ministry of Health; Genetics, Testing and Gene Patenting: Charting New Territory in Healthcare. 2002 http://www.health.gov.on.ca/english/public/pub/ministry_reports/geneticsrep02/report_e.pdf.
- 167.Organization for Economic Co-operation and Development. Genetic Inventions, Intellectual Property Rights & Licensing Practices: Evidence and Policies. Paris: Organization for Economic Co-operation and Development; 2002. http://www.oecd.org/dataoecd/42/21/2491084.pdf. [Google Scholar]
- 168.Oxford Gene Tech, Ltd. v. Affymetrix, Inc. Case No. 99-CV-348 (D. Del. 2000) [Google Scholar]
- 169.Oxford Gene Technology, Ltd. v. Affymetrix, Inc. Reports of Patent Design and Trademark Cases. 2001;118(9):310–328. [Google Scholar]
- 170.Paradise J, Andrews L, Holbrook T. Patents on human genes: an analysis of scope and claims. Science. 2005;307(5715):1566–1567. doi: 10.1126/science.1105162. [DOI] [PubMed] [Google Scholar]
- 171.Parahow L. Ariad Pharmaceuticals, Inc. et al. v. Eli Lilly and Co., Case No. 02-CV-11280 (D. Mass 2009) Brief of Amici Curiae, the Regents of the University of California, Wisconsin Alumni Research Foundation, The University of Texas System, University of Rochester, Rensselaer Polytechnic Institute, STC.UNM, The Research Foundation of State University of New York, NDSU Research Foundation, Research Corporation Technologies, Inc., En Banc Rehearing in Support of Affirmance of Judgment. [Google Scholar]
- 172.Parsons D. MS thesis. Durham, North Carolina: Duke Univ.; 2007. Seminal genomic technologies: Illumina, Inc., and high-throughput SNP genotyping BeadArray technology, a case study. http://www.genome.duke.edu/centers/cpg/documents/project1/bead-array/DBPARSONS%20MS%20THESIS%20-%20ILLUMINA%20FINAL.pdf. [Google Scholar]
- 173.Parthasarathy S. Building Genetic Medicine: Breast Cancer, Technology, and the Comparative Politics of Health Care. Cambridge, MA: MIT Press; 2007. [Google Scholar]
- 174.Pollack A. Amgen wins patent battle over Roche’s anemia drug. New York Times. 2007 Oct 24;:C2. [Google Scholar]
- 175.Powell A, Chandrasekharan S, Cook-Deegan R. Spinocerebellar ataxia: patient and health professional perspectives on whether and how patents affect access to clinical genetic testing. Genet. Med. 2010;12(4):S83–S110. doi: 10.1097/GIM.0b013e3181d67e44. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Press E, Washburn J. The kept university. Atl. Mon. 2000;285(3):39–54. [Google Scholar]
- 177.Prometheus Laboratories, Inc. v. Mayo Collaborative Services and Mayo Clinic Rochester. 92 U.S.P.Q.2d 1075 (Fed. Cir. 2009) [Google Scholar]
- 178.Amgen Inc. Q3 ‘09 earnings call. 2009 http://phx.corporate-ir.net/External.File?item=UGFyZW50SUQ9MzU1Mzk4fENoaWxkSUQ9MzQ2MzMyfFR5cGU9MQ==&t=1.
- 179.Rabinow P. Making PCR: A Story of Biotechnology. Chicago: Univ. of Chicago Press; 1996. [Google Scholar]
- 180.Rai A. Critical commentary on “open source” in the life sciences. 2009:213–218. See Ref. 96. [Google Scholar]
- 181.Splicing Life: The Social and Ethical Issues of Genetic Engineering with Human Beings. Washington, DC: US Government Printing Office; Randall C (National Council of Churches), Mandelbaum B (Synagogue Council of America), Kelly T (US Catholic Conference). 1982. Letter to President Jimmy Carter. In President’s Commission for the Study of Ethical Problems in Medicine and Biomedical and Behavioral Research; pp. 95–96. http://bioethics.georgetown.edu/documents/pcemr/splicinglife.pdf. [Google Scholar]
- 182.Reagan R. Public Papers of the Presidents of the United States: Ronald Reagan: 1983. vol. 1. Washington, DC: US Government Printing Office; 1983. Memorandum on government patent policy from February 18, 1983; p. 248. [Google Scholar]
- 183.Regents of the Univ. of Cal. v. Eli Lilly and Co. 39 U.S.P.Q.2d 1225 (S.D. Ind. 1995) [Google Scholar]
- 184.Regents of the Univ. of Cal. v. Eli Lilly and Co. 119 F.3d 1559 (Fed. Cir. 1997) [Google Scholar]
- 185.Reimers N. Tiger by the tail. CHEMTECH. 1987;17(8):464–471. [Google Scholar]
- 186.Reimers N. Stanford’s Office of Technology Licensing and the Cohen/Boyer cloning patents: an oral history. University of California, Berkeley: Regional Oral History Office, Bancroft Library; 2009. http://content.cdlib.org/view?docId=kt4b69n6sc&brand=calisphere&doc.view=entire_text. [Google Scholar]
- 187.Roberts L. Genome patent fight erupts. Science. 1991;254(5029):184–186. doi: 10.1126/science.254.5029.184. [DOI] [PubMed] [Google Scholar]
- 188.Robertson D. EST patent granted for human kinase homologs. Nat. Biotechnol. 1999;17(2):126. doi: 10.1038/6119. [DOI] [PubMed] [Google Scholar]
- 189.Russert B. Purdue University sequencing technology. Duke University, Durham, North Carolina: Center for Genome Ethics, Law and Policy; 2005. http://www.genomics.duke.edu/centers/cpg/project1/research-data/ [Google Scholar]
- 190.Secretary’s Advisory Committee on Genetics, Health, and Society. Washington, DC: US Department of Health and Human Services; Twentieth meeting of the Secretary’s Advisory Committee on Genetics, Health, and Society. Meeting proceedings. 2009 http://oba.od.nih.gov/oba/SACGHS/meetings/October2009/1009SACG-v2.pdf.
- 190A.Secretary’s Advisory Committee on Genetics, Health, and Society. Revised Draft Report on Gene Patents and Licensing Practices and Their Impact on Patient Access to Genetic Tests. Washington, DC: US Department of Health and Human Services; 2010. [Google Scholar]
- 191.Sell S. Industry strategies for intellectual property property and trade: the quest for TRIPS and post-TRIPS strategies. Cardozo J. Off. Intl. & Comp. Law. 2002;10:79–107. [Google Scholar]
- 192.Sell S. Private Power, Public law: The Globalization of Intellectual Property Rights. Cambridge, UK: Cambridge University Press; 2003. [Google Scholar]
- 193.Shiu C. Of mice and men: why an anticommons has not emerged in the biotechnology realm. Tex. Intell. Prop. L. J. 2009;7(3):413–456. [Google Scholar]
- 194.Shreeve J. The Genome War. New York: Ballantine Books; 2004. [Google Scholar]
- 195.Skeehan K, Heaney C, Cook-Deegan R. Impact of gene patents and licensing practices on access to genetic testing for Alzheimer’s disease. Genet. Med. 2010;12(4):S71–S82. doi: 10.1097/GIM.0b013e3181d5a68e. (First published 2009. Case study for the Secretary’s Advisory Committee on Genetics, Health, and Society, US Department of Health and Human Services, Washington, DC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Smith L, Sanders J, Kaiser R, Hughes P, Dodd C, et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321(6071):674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 197.Stevens AJ. The enactment of Bayh-Dole. J. Technol. Transf. 2004;29(1):93–99. [Google Scholar]
- 198.Stokes D. Pasteur’s Quadrant: Basic Science and Technological Innovation. Washington, DC: Brookings Institution Press; 1997. [Google Scholar]
- 199.Tait N. Ministers back EU patent reform plan. Financial Times. 2009 Dec. 4 http://www.ft.com/cms/s/0/193eccf2-e0eb-11de-9f58--00144feab49a.html.
- 200.Technology briefing. Biotechnology: Affymetrix and Hyseq resolve dispute. New York Times. 2001 Oct 26;:C5. [Google Scholar]
- 201.Ts’o P, Carter WA. No. 4,024,222. US Patent. 1977
- 202.UK Public Health Genetics Unit. Intellectual Property Rights and Genetics. Cambridge, United Kingdom: UK Public Health Genetics Unit; 2003. [Google Scholar]
- 203.US Const. Art. 1, Sec. 8, Clause 8
- 204.US House of Representatives. Introduced by Representative L Rivers with Representative D Weldon. Washington, DC: US House of Representatives; 2002. Genomic Research and Diagnostic Accessibility Act of 2002. [Google Scholar]
- 205.US House of Representatives. Introduced by Representative X Becerra with Representative D Weldon. Washington, DC: US House of Representatives; 2007. Genomic research and accessibility act. [Google Scholar]
- 206.US Senate. Proposed amendment to H.R. 2507, 102nd Congress. Proposed by Senator M Hatfield. Reprinted in US Senate. 1992. The genome project: the ethics issues of gene patenting. 1992:185–192. Serial No. J-102--83. Senate Hearing 102--1134 before the Subcommittee on Patents, Copyrights and Trademarks of the Committee on the Judiciary, US Senate, Washington, DC. [Google Scholar]
- 207.van Overwalle G. The Belgian compulsory license for public health [translation and summary of Debrulle J, De Cort L, Petit M. La, license obligatoire belge pour raisons de santé publique] 2007:199–209. See also Ref. 89. [Google Scholar]
- 208.van Overwalle G. Gene Patents and Collaborative Licensing Models: Patent Pools, Clearinghouses, Open Source Models and Liability Regimes. Cambridge, UK: Cambridge Univ. Press; 2009. [Google Scholar]
- 209.van Zimmeren E, Requena G. Ex-officio licensing in the medical sector: the French model. 2007:123–147. See ref. 89. [Google Scholar]
- 210.Varmus H. NIH Letter to Assistant Secretary of Commerce and Commissioner of Patents and Trademarks. Mar. 21, 1997. 1997 http://www.ott.nih.gov/policy/Archives.aspx.
- 211.Venter JC. A Life Decoded: My Genome: My Life. New York: Viking; 2007. [Google Scholar]
- 212.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 213.Venter JC, et al. No. 07/716831. US Patent App. 1991
- 214.Verbeure B. Patent pooling for gene-based diagnostic testing. Conceptual framework. 2009:3–32. See. Ref. 93. [Google Scholar]
- 216.Wade N. Scientist’s plan: map all DNA within 3 years. New York Times. 1998 May 10;:A1. [Google Scholar]
- 217.Wadman M. US court tests the breadth of patent protection on proteins. Nature. 2006;404(6778):532. doi: 10.1038/35007218. [DOI] [PubMed] [Google Scholar]
- 218.Wadman M. Anaemic outlook for Amgen. Nature. 2007;447(7147):899. doi: 10.1038/447899a. [DOI] [PubMed] [Google Scholar]
- 219.Walsh JP, Cho C, Cohen WM. Patents, Material Transfers and Access to Research Inputs in Biomedical Research: Final Report to the National Academy of Sciences’ Committee on Intellectual Property Rights in Genomic and Protein-Related Inventions. Washington, DC: National Academy of Sciences; 2005. [Google Scholar]
- 220.Walsh JP, Cho C, Cohen WM. View from the bench: patents and material transfers. Science. 2005;309(5743):2002–2003. doi: 10.1126/science.1115813. [DOI] [PubMed] [Google Scholar]
- 221.Washburn J. University, Inc.: The Corporate Corruption of Higher Education. New York: Basic Books; 2006. [Google Scholar]
- 222.Williamson AR. The Merck gene index project. Drug Discov. Today. 1999;4(3):115–122. doi: 10.1016/s1359-6446(99)01303-3. [DOI] [PubMed] [Google Scholar]
- 223.Williamson AR. Gene patents: Socially acceptable monopolies or an unnecessary hindrance to research? Trends Genet. 2001;17:670–673. doi: 10.1016/s0168-9525(01)02491-x. [DOI] [PubMed] [Google Scholar]
- 224.World Health Organization. Geneva: World Health Organization; Genetics, Genomics and the Patenting of DNA: Review of Potential Implications for Health in Developing Countries. 2005 http://www.who.int/genomics/patentingDNA/en/
- 225.Yamanaka K. A nail in the coffin for DNA sequence patents? Nat. Biotechnol. 2008;26:1085–1086. doi: 10.1038/nbt1008-1085. [DOI] [PubMed] [Google Scholar]
- 226.Zinner DE, Bjankovic D, Clarridge B, Blumenthal D, Campbell EG. Participation of academic scientists in relationships with industry. Health Affair. 2009;28(6):1814–1825. doi: 10.1377/hlthaff.28.6.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.35 U.S.C. 2006 http://www.gpoaccess.gov/uscode.
- 228.35 U.S.C. 200–212. 2006 http://www.gpoaccess.gov/uscode/
- 229.37 C.F.R. 401.1–401.17. 2008 http://www.gpoaccess.gov/cfr/