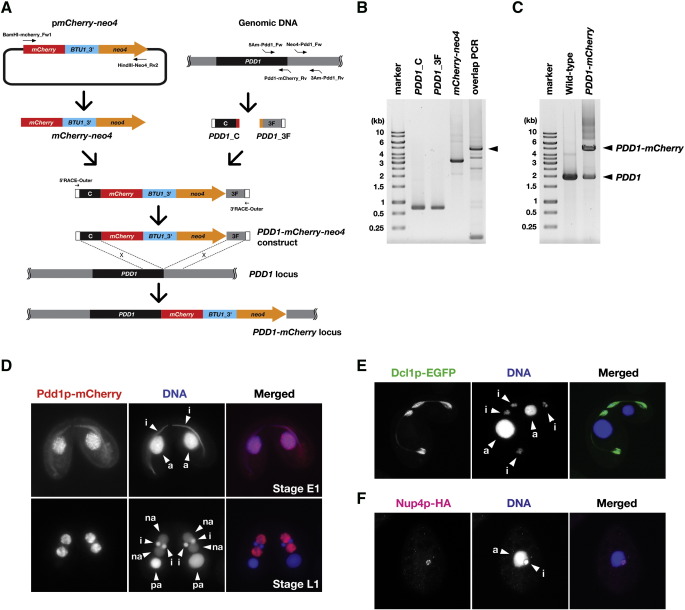
Fig. 3.
PCR-based C-terminal epitope tagging. (A) Production scheme for C-terminal mCherry tagging construct for PDD1 gene. See text for details. (B) Production of C-terminal mCherry tagging construct for PDD1 gene. The PCR products of PDD1_C, PDD1_F, mCherry-neo4 and PDD1-mCherry-neo4 (overlapping PCR) were analyzed. The position of PDD1-mCherry-neo4 construct is marked with an arrowhead. (C) PCR analysis of a PDD1-mCherry strain. Genomic DNA from both the wild-type B2086 and the PDD1-mCherry strains were amplified by PCR using the primers 5Am-Pdd1_Fw and 3Am-Pdd1_Rv. (D) Pdd1p-mCherry localization in Tetrahymena. The PDD1-mCherry strain shown in (C) was mated to a wild-type strain. Localization of Pdd1p-mCherry at conjugation stages E1 (top) and L1 (bottom) was observed using fluorescent microscopy (red). For reference, see Aronica et al. (2008) for conjugation stages. DNA was counterstained by DAPI (blue). a: macronucleus, i: micronucleus, na: new macronucleus, pa: parental macronucleus. (E) Dcl1p-EGFP localization in Tetrahymena. A DCL1-EGFP strain was mated to a wild-type strain and localization of Dcl1p-EGFP at conjugation stage E2 was observed under fluorescent microscope (green). DNA was counterstained by DAPI (blue). a: macronucleus, i: micronucleus. (F) Nup4p-HA localization in a vegetative Tetrahymena cell. Nup4p-HA was localized using an anti-HA antibody (magenta). DNA was counter-stained by DAPI (blue). The macronuclei and micronuclei are marked by “a” and “i”, respectively.