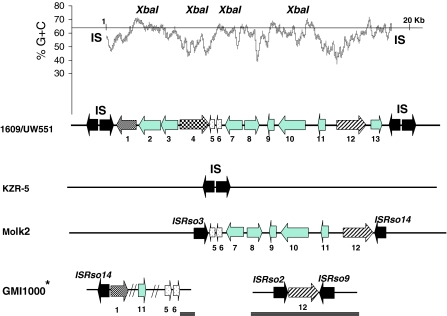
Fig. 3.
The PGI-1 region in strains 1609 and UW551, the deletion in strain KZR-5 and the related regions in strains Molk2 and GMI1000. The average G+C content of PGI-1 is shown over 200 bp intervals (upper graph). The line indicates the average G+C % over the whole genome (UW551; 64%). The location of four XbaI sites (positions 3,614, 5,330, 8,156 and 13,293) is also indicated. Positions 1 and 20 Kb correspond to position 243,626 (1) and 225,013 (20 Kb) of strain 1609. The location of the IS elements and the PGI-1 region (in between the IS blocks) in strains 1609 and UW551 are indicated (second graph). The organization of this region is, based on PCR and Southern hybridization results, the same in strain 715. Numbers below arrows indicate ORF numbers as in Table 4. ORF 1 through 3 encode hypothetical proteins, ORF 4 encodes a RelA/SpoT domain protein, ORF 5 a site specific integrase/recombinase, ORF 6 a bacteriophage related protein, ORF 7 a drug metabolite transporter (DMT) protein, ORF 8 a transcriptional regulator protein, ORF 9 through 11 hypothetical proteins, ORF 12 a cellobiohydrolase and ORF 13 a hypothetical protein. The organization of the relevant regions in strains KZR-5, Molk2 and GMI1000 are also shown. * ORF 1, 11, 5 and 6 localize on the chromosome, ORF 12 localizes on the megaplasmid.  Alternative codon usage region (ACUR)
Alternative codon usage region (ACUR)