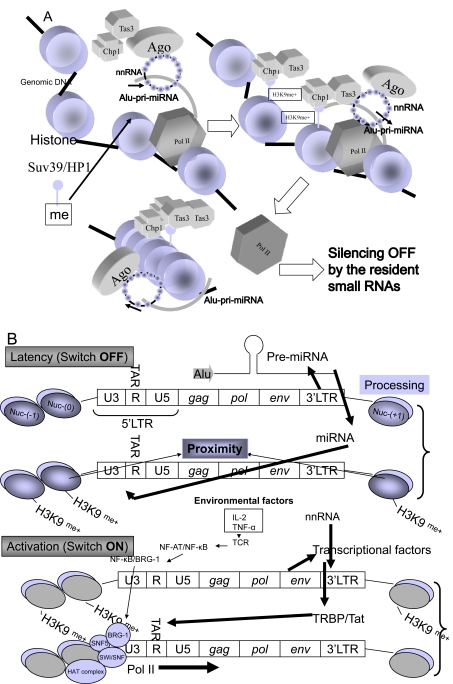
Fig. (3).
A model of epigetic on/off switching mechanism of transposable miRNAs. (A) Epigenetic silencing off by the resident small RNAs is shown. The function of nnRNA is similar to that of siRNA. Alu-pri-miRNAs as the TE could be targeted by nnRNA during transcription epigenetically. This process induces complete silencing off, that starts translation of mRNAs for differentiation of ESCs. The Ago/nnRNA complex circulized as a Buddhist rosary (‘Jyuzu’ in Japanease) and the ring of nnRNA may epigentically lock Alu-pri-miRNA transcription by methylation of histone by Clr4-like human methyltransferase (HMT) enzyme, Suv39 associated with Swi6/HP1 and chromo domain-containing protein 1 (Chp1) plus RNA-induced transcriptional silencing complex protein 3 (Tas3) binding to H3K9me+ histone following CpG methylation by DNA methyltransferase (DNMT) [111]. (B) The HIV-1 latency model. The Weinberg and Morris model [97] is shown with minor modifications. The HIV-1 3’LTR U3 region contains at least three genomic miRNA genes: MIR#4, MIRN367 and MIRH1. The 5’LTR/gag/pol/env region may be corresponding to Alu TE. These genomic miRNAs yeild silencing of HIV-1 proviral expression (Switch OFF). Environmental factors, such as the resident miRNA in ESMV, IL-2, X-ray, dietary food etc. affect the profile of miRNAs in cells, nnRNA from digestion of the resident miRNAs, retroposable miRNAs or mobile miRNAs completely stopped expression of Alu-pri-miRNA described in (A). This is the mechanism of latency. Dominant transcriptional factor induces HIV-1 mRNA transcription (Switch ON). In this state, iPS cells may be produced; however, since there is no step of complete silencing on, iPS cells may occasionally change into tumour cells. Therefore, the differentiation set of the resident miRNAs should be completely switched off to reprogram the somatic cell.