Abstract
A persistent problem in heterologous protein production is insolubility of the target protein when expressed to high level in the host cell. A widely employed strategy for overcoming this problem is the use of fusion tags. The best fusion tags promote solubility, may function as purification handles and either do not interfere with downstream applications or may be removed from the passenger protein preparation. A novel fusion tag is identified that meets these criteria. This fusion tag is a monomeric mutant of the Ocr protein (0.3 gene product) of bacteriophage T7. This fusion tag displays solubilizing activity with a variety of different passenger proteins. We show that it may be used as a purification handle similar to other fusion tags. Its small size and compact structure are compatible with its use in downstream applications of the passenger protein or it may be removed and purified away from the passenger protein. The use of monomeric Ocr (Mocr) as a complement to other fusion tags such as maltose-binding protein will provide greater flexibility in protein production and processing for a wide variety of protein applications.
Keywords: Fusion protein, Solubility enhancement, Purification handle, Passenger protein
Due to its low cost, compatibility with automation and ease of scale-up, Escherichia coli remains the most widely used host for high-throughput protein production [1–3]. A major hurdle for heterologous protein production in E. coli is the formation of insoluble aggregates. This problem is commonly addressed through the use of fusion tags to enhance solubility [4–6]. Comparative studies of the effectiveness of fusion tags have shown the maltose-binding protein (MBP)1 to be one of the best at solubilizing passenger proteins [7,8]. The properties that make a fusion tag capable of enhancing solubility are not fully understood, although the acidity of the fusion tag is often correlated with this capability [9,10]. Due to MBP’s solubilizing capability and its affinity for amylose, which allow it to be used as an affinity handle, vectors containing MBP fusion tags have been developed for use in high-throughput cloning and expression [11].
Although MBP is quite effective in solubilizing its passenger proteins during expression, a number of problems have been identified with its use that occurs during purification and processing of the fusion. MBP fusions do not always bind to amylose resin and so a His tag is commonly added to facilitate affinity purification using a metal chelating resin [12]. Many proteins that are soluble when fused to MBP have been observed to precipitate when the MBP-fusion is cleaved [11]. Additionally, the incomplete removal of MBP from the passenger protein after cleavage of the fusion [11] may interfere with downstream applications such as NMR or crystallization.
We sought to develop a new fusion partner with solubilizing capabilities similar to MBP while avoiding or reducing the problems found in MBP fusion purification and processing. The bacteriophage T7 0.3 protein (Ocr for ability to overcome classical restriction) is a 13.8 kDa, highly charged, very acidic (pI = 3.8) protein [13]. It has an efficiently translated transcript and is tolerated at high levels in the cell. There are no cysteines in the Ocr protein [13]. The protein is completely soluble even when expressed to high level and is soluble in 95% ethanol [14]. The Ocr protein may be separated to high levels of purity from E. coli using DEAE resins [14]. These properties suggested it could make a robust fusion partner to promote target protein production and solubility.
In its native state, the Ocr protein forms a dimer, which could potentially foster aggregation of a fused partner. The crystal structure of the Ocr dimer revealed a small, hydrophobic subunit interface [15]. Here we report two amino acid substitutions that disrupt dimer formation and that stabilize the monomeric form of the protein, which we call Mocr, for monomeric Ocr. We further characterize Mocr as a fusion partner that displays solubilizing activity with problematic passenger proteins.
Materials and methods
Construction and cloning of mocr
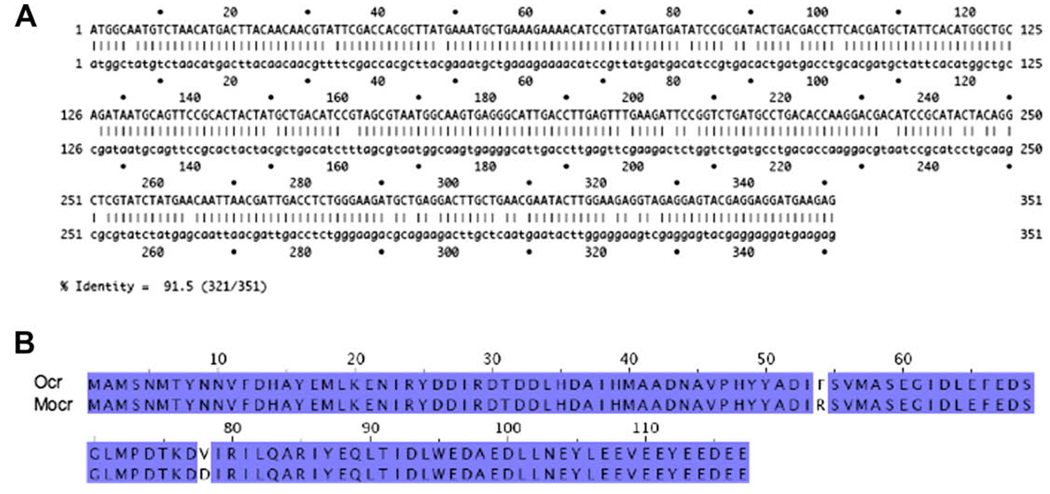
A synthetic sequence of the ocr gene was created to yield oligos with minimal dimerization potential and self-annealing. The only purpose of the silent mutations contained in the synthetic sequence was to improve the efficiency of the synthetic gene construction. This sequence also incorporated mutations that change the amino acid sequence. Codon 53 has been changed from TTT to CGT and codon 77 has been changed from GTA to GAC (Fig. 1A). These codon changes gave rise to the following amino acid changes: F53R and V77D. The amino acid sequence of Mocr is shown in Fig. 1B.
Fig. 1.
(A) Strider alignment of the DNA sequence of the synthetic mocr gene (top) and the native ocr gene (bottom). The silent mutations in the synthetic sequence were introduced to reduce self-annealing or dimerization in the oligos used to construct the synthetic gene. (B) A Jalview (www.jalview.org) alignment of the Ocr amino acid sequence (top) with the mutant Ocr (Mocr) amino acid sequence (bottom) is shown. Identical amino acids are highlighted. The Mocr sequence contains two mutations, F53R and V77D. These were made to disrupt the dimer interface.
The gene was then constructed through a combination of sequential oligo pair annealing and ligation and PCR amplification. For sequential oligo pair annealing and ligation, the oligos were resuspended in water to 20 µM. Each oligo was the phosphorylated in a 20 µl reaction containing 2 µl ATP (10 mM), 2 µl 10× ligation buffer (NEB), 12.5 µl of the oligo, 2.5 µl H2O and 1 µl of polynucleotide kinase. Reactions were incubated at 37 °C for 1 h, then at 95 °C for 10 min. Complementary oligo pairs were mixed and slow cooled to 37 °C to create double-stranded fragments. Adjacent fragments were then mixed in equal volume and slow cooled to 4 °C. At 4 °C additional ATP was added along with 2 U of T4 DNA ligase and incubation was continued at 4 °C for 1 h. Mixing of adjacent fragments at 37 °C followed by slow cooling and ligation was continued until all the fragments had been mixed. The resulting fragment was then PCR amplified using outside primers to add a BglII restriction site to the 5′-end and a KpnI site to the 3′-end. The resulting fragment was gel purified and cloned into pMCSG7 [16] digested with BglII and KpnI.
Cloning wild-type ocr
Bacteriophage T7 DNA was obtained from ATCC (BAA-1025-B2). Phage were produced by transfection of strain HMS174 (ATCC 47011) with resuspended DNA. The cells were grown until lysis. This broth was used to infect two 5 ml cultures of HMS174. After lysis the cell debris was spun out. The phage was precipitated by the addition of PEG with incubation at 4 °C overnight followed by centrifugation. The pellets were resuspended in a total of 1 ml of PBS. A 200 µl aliquot was extracted with phenol/chloroform and the DNA was then precipitated by addition of ammonium acetate and ethanol. This DNA was used as PCR template in reactions using outside primers to add a BglII restriction site to the 5′-end and a KpnI site to the 3′-end. The resulting fragment was gel purified and ligated with pMCSG7. Positive clones were identified by PCR and confirmed by DNA sequencing.
Gel filtration of Ocr and Mocr
Cultures (250 ml) in terrific broth (TB: 6 g tryptone, 12 g yeast extract, 4% (20 ml) glycerol, 1.15 g KH2PO4, 6.25 g K2HPO4 in 500 ml distilled water) were grown in 1 L flasks at 37 °C, 250 rpm to an OD at wavelength 600 of approximately 1. The temperature was reduced to 20 °C and after equilibration at this temperature for 1 h the cultures were induced by addition of 200 µM IPTG. Incubation was continued at 20 °C overnight (18 h). Cultures were centrifuged and pellets were frozen at −80 °C. Cell pellets (5–6 g) were resuspended in 40 ml PBS with 0.1 mg/ml lysozyme and benzonase and lysed by sonication. Lysate was centrifuged at 20,000g for 1 h. Soluble fraction was batch bound overnight to 2 ml Ni-NTA agarose from Qiagen. Resin was washed with 20 mM imidazole in PBS and eluted off with 250 mM imidazole in PBS. The 10–15 ml eluate was dialyzed in 50 mM Tris pH 8.0, 150 mM NaCl, 0.1 mM EDTA, and 1 mM DTT. Gel filtration was performed using a 120 ml Superdex 75 column on an Akta Explorer FPLC. The running buffer composition was the same as the dialysis buffer.
DEAE chromatography
Soluble fraction was batch bound to 2 ml DEAE resin for 2 h. DEAE chromatography was performed using DEAE Ceramic HyperD F resin from Pall Life Sciences. The resin was washed with 50 mM sodium phosphate and varying concentrations of NH4Cl, from 50 to 400 mM, and then eluted with 50 mM sodium phosphate and varying concentrations of NH4Cl, from 400 mM to 1 M.
High-throughput construct design, cloning, expression and purification
Design
Protein sequences were analyzed for ordered and disordered regions using server based programs: Foldindex (http://bioportal.weizmann.ac.il/fldbin/findex) and DisEMBL (http://dis.embl.de/). Secondary structure was predicted using Jpred (http://www.compbio.dundee.ac.uk/~www-jpred/submit.html). This information was overlaid with prior biochemical and functional domain information to design constructs. A full-length native construct was always included as a standard for comparison for protein production level of the various constructs. The other constructs were variations on the gene sequence. The different start and stop sites designed for the gene of interest were combined with each other to create a matrix of different truncated forms of the protein which were tested for soluble protein production in E. coli.
Oligos for PCR construction were designed using Clone-manager (Scientific and Educational Software). The following overlaps for ligation-independent cloning (LIC) were added to each oligo: coding strand 5′-TACTTCCAATCCAATGCN and non-coding strand 5′-TTATCCACTTCCAATG (TTA or CTA). For the coding strand the last three bases are an alanine codon. N is any base. An A or T is found in this position in the highest use codons for alanine in E. coli.
PCR construction and cloning
The constructs were produced in 50 µl PCRs to which was added 5 µl 10× buffer, 1 µl of 50 mM MgSO4, 1.5 µl of 10 mM dNTP mix, 5 µl of PCR Enhancer Solution (Invitrogen), 0.5 µl of 2.5 U/µl Platinum Pfx DNA polymerase (Invitrogen) and 0.5 µl of miniprep plasmid DNA template. The program used begins with 2.5 min at 94 °C followed by 30 cycles of 30 s at 94 °C, 30 s at 50 °C and 2 min at 68 °C. The final step is an additional 3.5 min at 68 °C and a reduction to 4 °C. The reactions were cleaned up using MinElute 96 UF PCR purification kits by Qiagen. These were run on a Biomek FX liquid handler.
The LIC vector was linearized with SspI (New England Biolabs) at 5 U/µg DNA incubated 1.5 h at 37 °C. The protein was removed using a Qiagen PCR spin kit. The linearized DNA was processed by T4 DNA polymerase to yield single-stranded ends for annealing. The 60 µl reaction contained 1.6–2.0 µg linear DNA, 6 µl 10× buffer, 3 µl 100 mM DTT, 2.4 µl 100 mM dGTP, 1.5 µl 2.5 U/µl T4 DNA polymerase (Novagen LIC qualified), 41.1 µl dH20, 6 µl Template (200–300 ng/µl). These were mixed on ice and then incubated in a PCR machine for 30 min at 22 °C, shifted to 75 °C for 20 min, and the temperature was reduced to 4 °C.
Constructs of the genes of interest were treated with T4 DNA polymerase, to prepare them for annealing, in 20 µl reactions using 5 µl of the cleaned up PCR as substrate. A 1 µl aliquot of 100 mM DTT, 0.8 µl of 100 mM dCTP, 2 µl 10× buffer and 0.5 µl of T4 DNA polymerase was added to each reaction. These were incubated as for the vector.
Annealing was carried out in 96-well plates. A 1 µl aliquot of vector was mixed with 2 µl of insert on ice. The plate was placed in a PCR machine and incubated for 10 min at 22 °C. A 1 µl aliquot of 25 mM EDTA was added to each reaction and incubation was continued for 5 min at 22 °C. The plate was chilled on ice and a 1 µl aliquot of each reaction was used to transform 20 µl of Z-competent XL1Blue cells. These were plated on LB agar 48-well Q-trays (Genetix) containing ampicillin for selection. Colonies were picked and grown in 1 ml of LB overnight in 96-well deep well blocks and then plasmid DNA was miniprepped from these cultures using Perfect Prep 96 Vac purification kits by Eppendorf on the Biomek FX. Positives clones were identified by PCR analysis.
Expression
A 1 µl aliquot of the miniprep DNA for each positive clone was used to transform competent Rosetta2 cells in 96-well plates. The recovered transformation mixes were used to inoculate 1 ml of LB containing ampicillin and chloramphenicol as selective agents. These were grown overnight at 37 °C and used to both create glycerol stocks and inoculate 0.5 ml of terrific broth in a 96-well deep well block. The TB block was grown at 37 °C and shaking at 400 RPM to an OD600 of approximately 1. The temperature was reduced to 20 °C and after equilibration at this temperature for 1 h the cultures were induced by addition of IPTG to the specified concentration. Incubation was continued at 20 °C overnight (18 h). Cultures were frozen at −80 °C.
High-throughput purification
Frozen culture blocks were thawed at 25 °C. A 1.26 g bottle of CelLytic Express (Sigma) was resuspended in 5 ml of PBS. A 50 µl aliquot of CelLytic was added to each well of the 96-well deep well block. These were incubated at 25 °C with shaking for 20 min for lysis. These were transferred to 1.1 ml Axygen minitubes and centrifuged for 10 min at 20,000g. The soluble fraction was transferred to a 96-well deep well block containing 120 µl of a slurry of 75% PBS and 25% Ni-NTA Agarose from Qiagen. Protein was allowed to batch bind to resin for 1 h at 4 °C. The purification was performed on a Biomek FX. The resin and soluble fraction were transferred to a Whatman 96-well 2 ml glass filled 10 µm polypropylene filter. The resin was washed six times by 400 µl of 20 mM imidazole in PBS. The protein was eluted by 250 mM imidazole in PBS.
TEV cleavage of the Mocr-Med15 fusion and purification by size exclusion chromatography
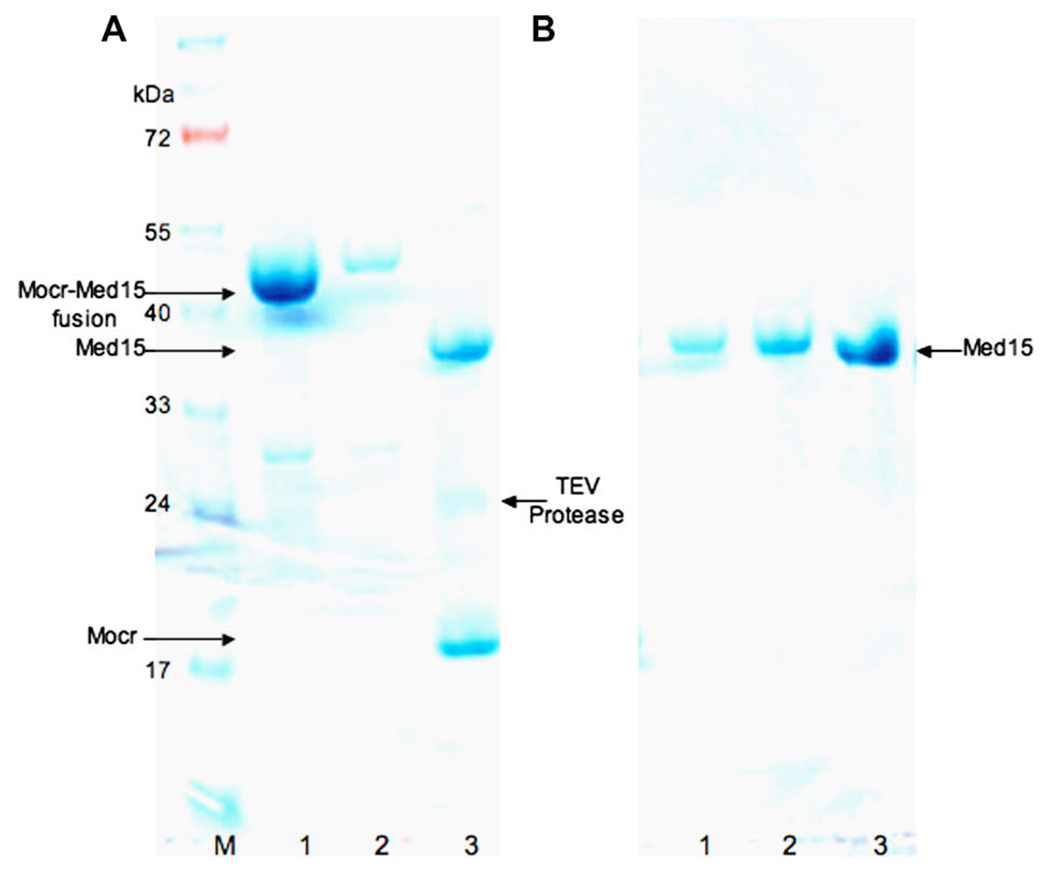
The soluble fraction from cell lysates was batch bound to 2 ml Ni-NTA resin. The fusion protein was eluted with three 2 ml aliquots of 100 mM Tris (pH 7.5), 100 mM NaCl, 10 mM β-mercatoethanol, 10% glycerol, and 300 mM imidazole. The three elutions were pooled and dialyzed in the presence of tobacco etch virus (TEV) protease overnight (18 h) at 4 °C in 50 mM Tris (pH 7.5), 100 mM NaCl, 10 mM β-mercatoethanol, and 10% glycerol. Typically the TEV protease was added at a ratio of roughly 1 to 20–25 mg of protease to milligrams of protein of interest using a 1 mg/ml stock of protease. For this particular experiment (Fig. 8) there was 50 mg of Mocr-Med15 fusion and 2 mg of TEV protease present. Cleaved protein was then subjected to size exclusion chromatography using a Superdex 75 column on an Akta Explorer FPLC. The Med15 containing fractions were pooled and concentrated using a Vivascience 30 K centrifugal filter to a final concentration of 10 mg/ml.
Fig. 8.
SDS–PAGE analysis of TEV cleavage of a Mocr-Med15 fusion protein. The soluble fraction from cell lysates was batch bound to 2 ml Ni-NTA resin. The fusion protein was eluted with three 2 ml aliquots of imidazole containing buffer. The three elutions were then pooled and dialyzed in the presence of TEV protease overnight to cleave the fusion. The Med15 portion of the fusion was then separated from Mocr by gel filtration using a Superdex 75 column. (A) Molecular weight marker (M), first imidazole elution from NiNTA (1) third imidazole elution from NiNTA (2) TEV cleavage of fusion (3). (B) Gel filtration fractions (1), (2), pooled fractions concentrated 5× (3). The gel was stained with Coomassie blue. The proteins are identified with arrows.
Sample analysis
All analysis of PCR and high-throughput protein expression samples was done using a Labchip90 instrument (Caliper). The protein obtained from DEAE-purification trials was analyzed by SDS–PAGE using 26 lane 4–20% Tris–Gly gels followed by staining with Bio-Safe Coomassie (BioRad).
Results
Monomerization of Ocr protein
Gene 0.3 or ocr, is the first gene found in the linear bacterio-phage T7 genome, and is also the first gene transcribed upon phage entry into the host cell. The protein encoded by ocr is an inhibitor of E. coli restriction enzymes [17]. Ocr protein inhibits E. coli type I restriction enzyme by forming an extended dimer that mimics B-form DNA [15]. The dimer binds the restriction enzyme in competition with the DNA substrate. The binding affinity between Ocr and EcoKI is extremely strong, with a dissociation constant estimated at 100 pM [18]. We sought to disrupt dimer formation to prevent fusion proteins from being bound by the restriction enzymes as well as to reduce the possibility that dimerization could promote formation of large aggregates when Ocr is fused to aggregation-prone proteins.
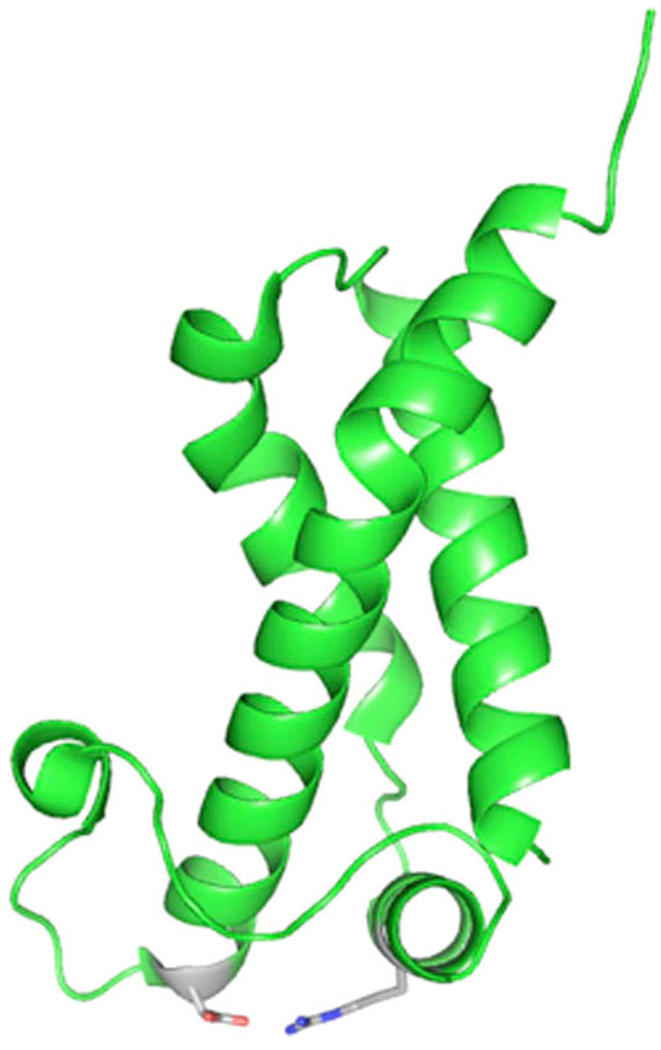
The small interface between the Ocr monomers is composed of nonbonded hydrophobic contacts and no hydrogen bonds [15]. Phe 53 of one monomer appears as a “ knob” that fits into a “hole” surrounded by Ala 50, Ser 54, Met 56, and Ala 57 of the other monomer. A second contact site occurs between Val 77 of the two monomers. To disrupt this interface with the smallest number of mutations, we substituted Phe53 with arginine. The charged amino acid should disfavor the insertion of the “knob” into the hydrophobic “hole”. Val 77 was substituted with aspartic acid to replace the van der Waals interaction with charge repulsion between the acidic side chains. The basic arginine and the aspartic acid were designed to be close enough to potentially establish a salt bridge, which may assist in stabilizing the monomeric structure (Fig. 2). A synthetic version of the ocr gene was designed to introduce these mutations (Fig. 1).
Fig. 2.
The crystal structure of Ocr deposited in the PDB (1S7Z.pdb) was used to produce this proposed structure. The amino acids highlighted are the aspartate (in red) that replaced the valine at position 77 and the arginine (blue) that replaced the phenylalanine at position 53. These are oriented to show how they could potentially form a salt bridge that could increase the stability of the monomeric form of Ocr. The predicted distance between these functional groups is 2.5 Å. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
The gene sequence shown in Fig. 1A was constructed and cloned in frame into vector pMCSG7 [16]. The resulting vector, pMocr (monomeric ocr), has a T7 promoter linked to a coding sequence for a His6-tag, a short spacer followed by the mocr sequence and a sequence coding for a TEV cleavage site at the end (Fig. 3). A ligation-independent cloning (LIC) site in the DNA encodes a portion of the tobacco etch virus (TEV) cleavage site. The native ocr gene was also inserted into the same sites in pMCSG7 for comparison with the mutated version.
Fig. 3.
The amino acid sequence of the synthetic Ocr as it appears in the pMCSG7 vector. The synthetic gene coding for putative monomeric Ocr was cloned into pMCSG7 from BglII to KpnI. This places the synthetic Ocr coding sequence downstream of the His6 tag. There is a short spacer sequence and then the Ocr coding sequence begins, shown in italics above. At the end of the Ocr sequence there is a glycine and then the Tobacco Etch Virus (TEV) protease recognition sequence (underlined). The pMCSG7 vector was designed so that the TEV recognition sequence overlaps the site of LIC annealing.
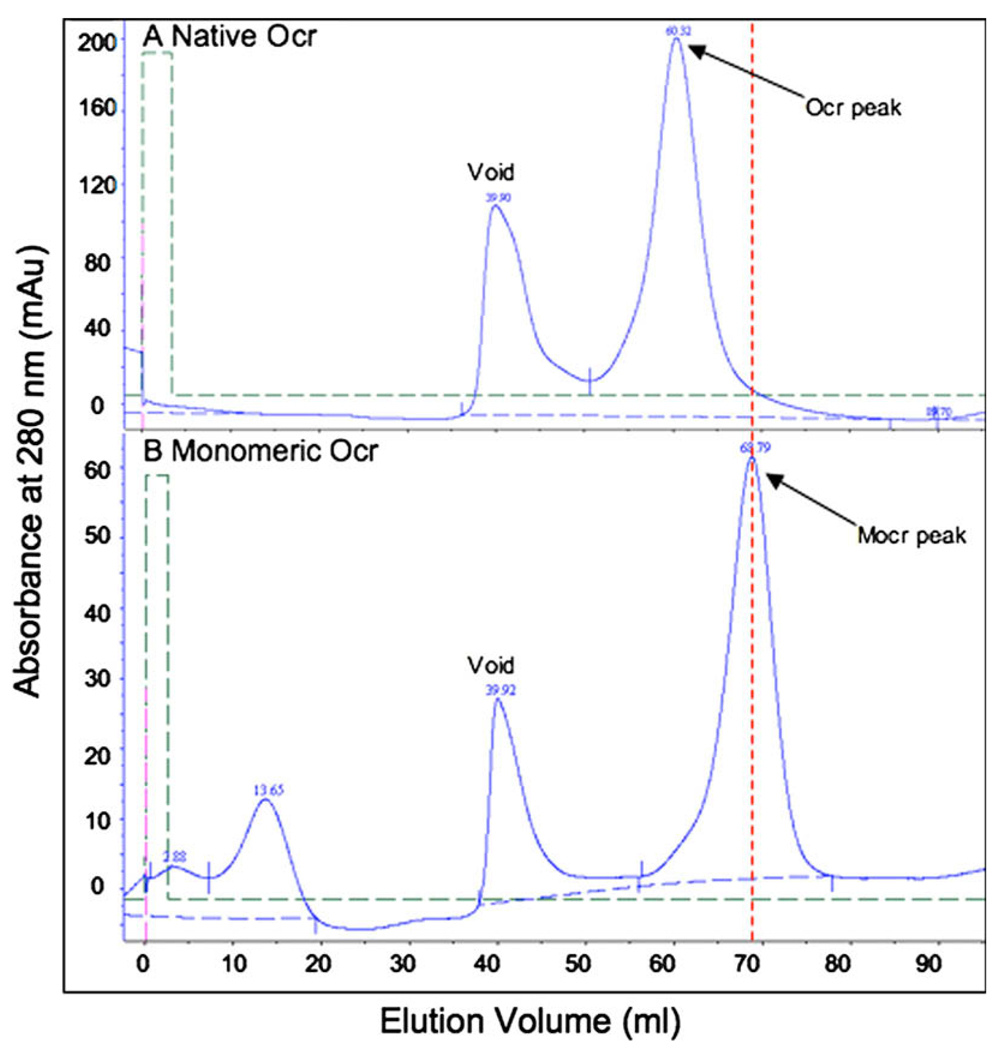
The two forms of Ocr were produced in BL21 (DE3) cells and purified by nickel chelate chromatography then subjected to size exclusion chromatography. Fig. 4A is the profile for size exclusion chromatography of the native Ocr protein, which should run as a dimer under non-denaturing conditions. Native Ocr elutes from the size exclusion column at a calculated molecular weight of 35 kDa. The predicted molecular weight for the native Ocr protein in pMCSG7 is 16.6 kDa, which would yield a dimer of 33.3 kDa. The native protein appears to be a dimer under these conditions. When Mocr protein is run on the same size exclusion resin under identical conditions the elution peak is shifted to a later volume (69 ml versus 60 ml, Fig. 4B). The Mocr protein elutes from the size exclusion column at a calculated molecular weight of 21 kDa. The predicted molecular weight for the Mocr protein as produced from the pMCSG7 vector is 16.7 kDa. This appears to be monomeric under these conditions. When the Mocr is cleaved from passenger protein using TEV protease during the purification of fusion proteins the released Mocr elutes from gel filtration at a calculated molecular weight between 16 and 20 kDa (data not shown). Thus, the Mocr protein reproducibly behaves as a monomer in diverse contexts.
Fig. 4.
SEC profiles for native (A) and monomeric (B) Ocr proteins. Samples were run on a 120 ml Superdex 75 column in 50 mM Tris (pH 8.0) with 150 mM NaCl. The chromatography profiles shown above plot the UV absorbance detected at 280 nm versus the volume of buffer (milliliters) run over the column.
Mocr fusion has solubilizing activity
We used the Mocr fusion with a variety of target proteins (A through D below) and compared its performance to several others fusion tags in use in our lab. The objectives for target protein production ranged from biochemical characterization to high-throughput screening for ligands to solving the structure by crystallography or NMR. Four representative target proteins were included in this study (Table 1).
Table 1.
Solubilization of passenger proteins by Mocr fusions
| Protein | Total constructs | Soluble constructs | ||||
|---|---|---|---|---|---|---|
| (Mol. wt. rangea) | His | GST | MBP | GB1 | Mocr | |
| Caveolin | 22 (7–20.5 kDa) | 0 | NDb | 6 | 0 | 5 |
| Acyl trans. | 32 (31–100 kDa) | 0 | ND | 17 | ND | 14 |
| MAV-1 | 24 (25–55 kDa) | 2 | ND | 24 | 19 | 18 |
| MNV | 20 (18–59 kDa) | 5 | 7 | 18 | ND | 20 |
The molecular weight range is of the constructs for each protein and only includes the passenger protein and not the fusion tag. The largest molecular weight corresponds to the full-length native form of the protein and the others represent a matrix of truncations at either or both ends of the protein.
Not done.
A. Caveolin is found in caveolae, vesicular invaginations of the plasma membrane. Caveolae function in vesicle trafficking, cholesterol homeostasis, signal transduction and tumor suppression. Caveolin has several distinct domains, an N-terminal intercellular domain, which has a phosphorylation site, an oligomerization domain and a transmembrane domain. Recombinant caveolin is produced primarily in eukaryotic cells although bacterial production of the N-terminal domain as a glutathione-S-transferase (GST) fusion protein has been reported [19]. The goal was to find stable, soluble forms of the caveolin protein as wild type and also as phosphorylation mutants and as mutants of the oligomerization domain. Most of the constructs were truncations although full-length versions were also tried.
B. Bryostatin is an anti-cancer compound synthesized by an uncultured symbiont of a marine invertebrate [20]. Characterizing the activities of the biosynthetic enzymes in the pathway of bryostatin may allow for the development of novel combinations of enzymes that would yield a wider variety of bryostatin analogues with different therapeutic properties. The full-length version of an acyl transferase from this pathway did not yield soluble protein when expressed in E. coli. The goal for this project was to obtain soluble protein for biochemical characterization and for structural studies.
C. The penton and fiber knob proteins of adenovirus are found at the vertices of the icosahedral capsid. These proteins are involved in binding cellular receptors including integrins, triggering internalization via endocytosis. The mouse adenovirus type 1 (MAV-1) fiber knob protein has a unique loop sequence not found in other homologs. To facilitate both biochemical and structural characterization, we designed a variety of constructs of each gene for expression in E. coli.
D. Noroviruses, members of the Caliciviridae family, infect primarily humans but also pigs, cattle and mice. Human noroviruses are the major cause of nonbacterial epidemic gastroenteritis worldwide resulting in substantial morbidity and economic loss but no drugs or vaccines are available for treatment. Virus-receptor interaction and virus entry have become attractive targets for anti-viral therapies. To facilitate this goal we designed constructs encoding full-length or truncated versions of the mouse norovirus (MNV) major capsid gene VP1 or its two domains (“shell” and “protruding” [P]) for expression in E. coli.
The different constructs for each gene were cloned into several fusion vectors and expression was carried out in a Rosetta BL21 (DE3) strain of E. coli. All the vectors were based on the pMCSG vectors [11,19] and have a sequence encoding a His6 tag followed by either a spacer or a fusion tag and then a TEV cleavage site encoding sequence adjacent to the gene of interest. The cultures were subjected to chemical lysis and the insoluble fractions were separated by centrifugation. The proteins of interest were purified by chromatography on nickel resin in a 96-well plate format. The eluates were analyzed for protein using a Caliper Labchip90. A protein was considered soluble if it was detected at >20 ng/µl by the Labchip90 analysis. This level of production would be roughly equivalent to 2 mg of purified protein per liter of culture.
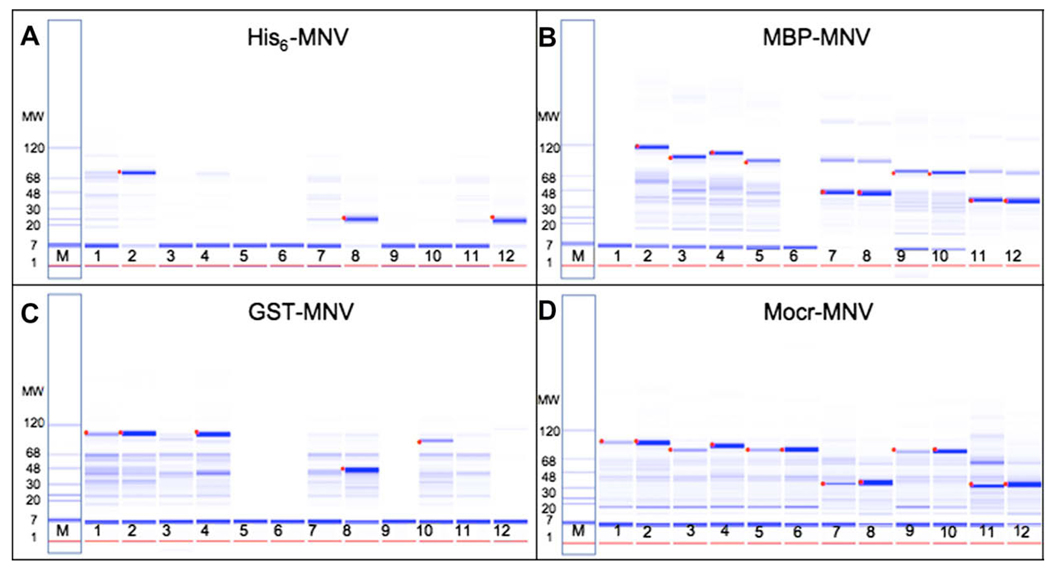
Only 7 of the 98 constructs were detectable in the soluble fraction when a His6 tag was used alone. For all protein targets, Mocr displayed solubilizing activity (Table 1) comparable to MBP, which is one of the most effective solubilizing fusion tags [7,8]. By comparison, fusions with GST or immunoglobulin-binding domain of streptococcal protein G (GB1) displayed lower efficiency in solubilizing the constructs. The data for a representative set of samples (MNV construct fusions) from the Labchip analysis of soluble protein in the eluate from the nickel resin purification step are shown in Fig. 5. The dot identifies the protein at the expected molecular weight for each construct. Although the pattern of solubilization between Mocr and MBP was similar, the yield for individual constructs varied among the fusion tags. In several of the MBP lanes [7,8,11,12] there appear to be multimers of the protein of interest forming. We occasionally observe this in samples of high protein concentration. The standard Labchip sample buffer does not contain a reducing agent and we believe these multimers are the result of disulfide bond formation between cysteines in the passenger portion of the fusion protein.
Fig. 5.
Labchip 90 analysis of MNV major capsid protein constructs for soluble expression in different fusion vectors. These are representative samples from the expression of MNV major capsid protein fusions. The data is shown as a gel representation similar to SDS–PAGE stained with coomassie blue. (A) His6 fusions; (B) MBP fusions; (C) GB1 fusions; and (D) Mocr fusions. The samples are the eluates from the nickel chelate chromatography step of high-throughput expression and purification as described in Methods and Materials. The first lane (labeled M) in each panel is a protein molecular weight ladder ranging from 1 to 120 kDa. Each lane (numbered 1–12) is the elution sample from expression of a different construct of the MNV major capsid protein gene and indicated fusion tag. The dot identifies the protein migrating at the expected molecular weight for each construct and fusion. The same constructs are shown for each fusion tag.
Mocr can function as a purification handle
The Ocr protein is not a standard affinity tag, however it has a strong interaction with DEAE-cellulose resin. In the initial reported purification [14] Ocr was bound to DEAE resin in the presence of 0.3 M NH4Cl, a condition at which few other proteins in the E. coli lysate bound to the resin. The Ocr protein was then eluted at 95% purity with 0.5–0.6 M NH4Cl.
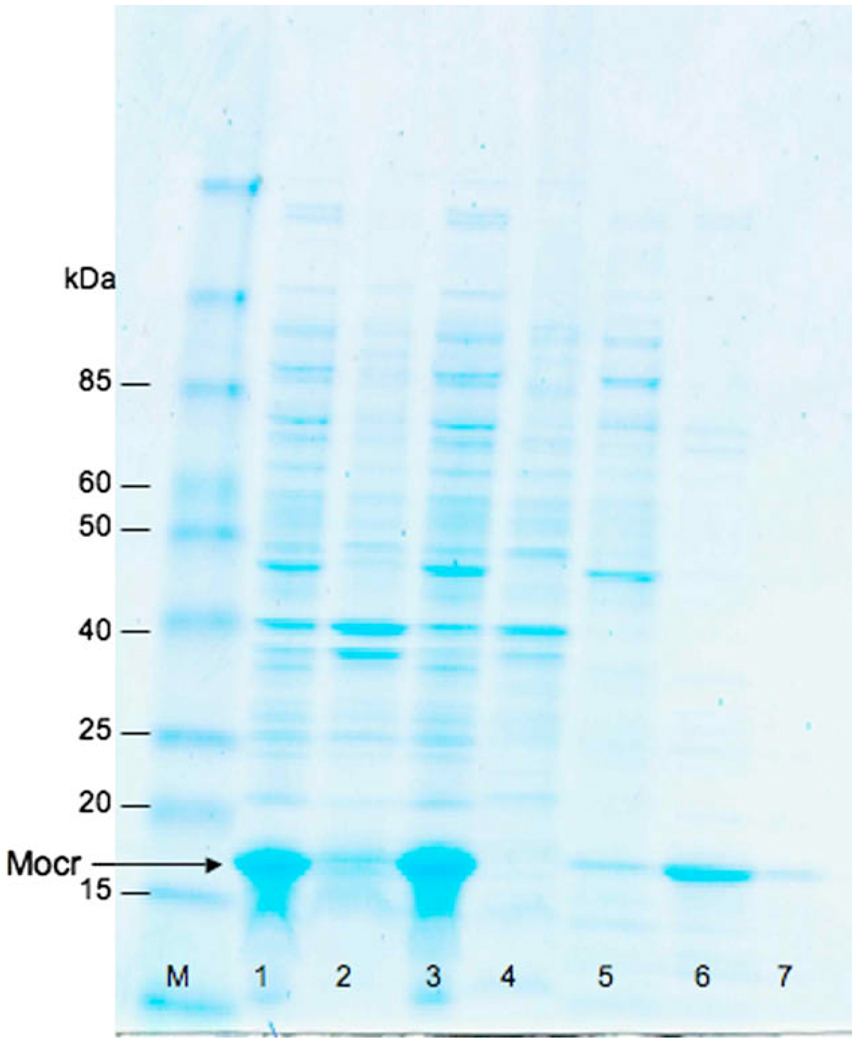
We examined the ability to purify Mocr as configured in our fusion vector from bacterial lysates using DEAE-cellulose. A matrix of wash and elution conditions was run to find the optimal combination for yield and purity. In these experiments, the Mocr encoded by our vector behaved similarly to the native Ocr during purification by DEAE-cellulose. Optimal conditions were slightly different than those described previously [14], with a 200 mM NH4Cl wash followed by a 600 mM NH4Cl elution yielding the best results. We repeated the purifications at a larger scale to confirm these results (Fig. 6). We estimate the purity of the eluted protein at greater than 80%. Treatment of the resin with SDS to remove any uneluted protein showed a small amount did remain bound after the elution (Fig. 6, lane 7).
Fig. 6.
SDS–PAGE analysis of wash and elution conditions for purification of Mocr protein on DEAE-cellulose resin. Molecular weight marker (M), whole cell extract (1), insoluble fraction (2), soluble fraction (3), flow-through (4), 200 mM NH4Cl wash (5), 600 mM NH4Cl elution (6), resin (7). The gel was stained with Coomassie blue. The position of Mocr protein is identified by the arrow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
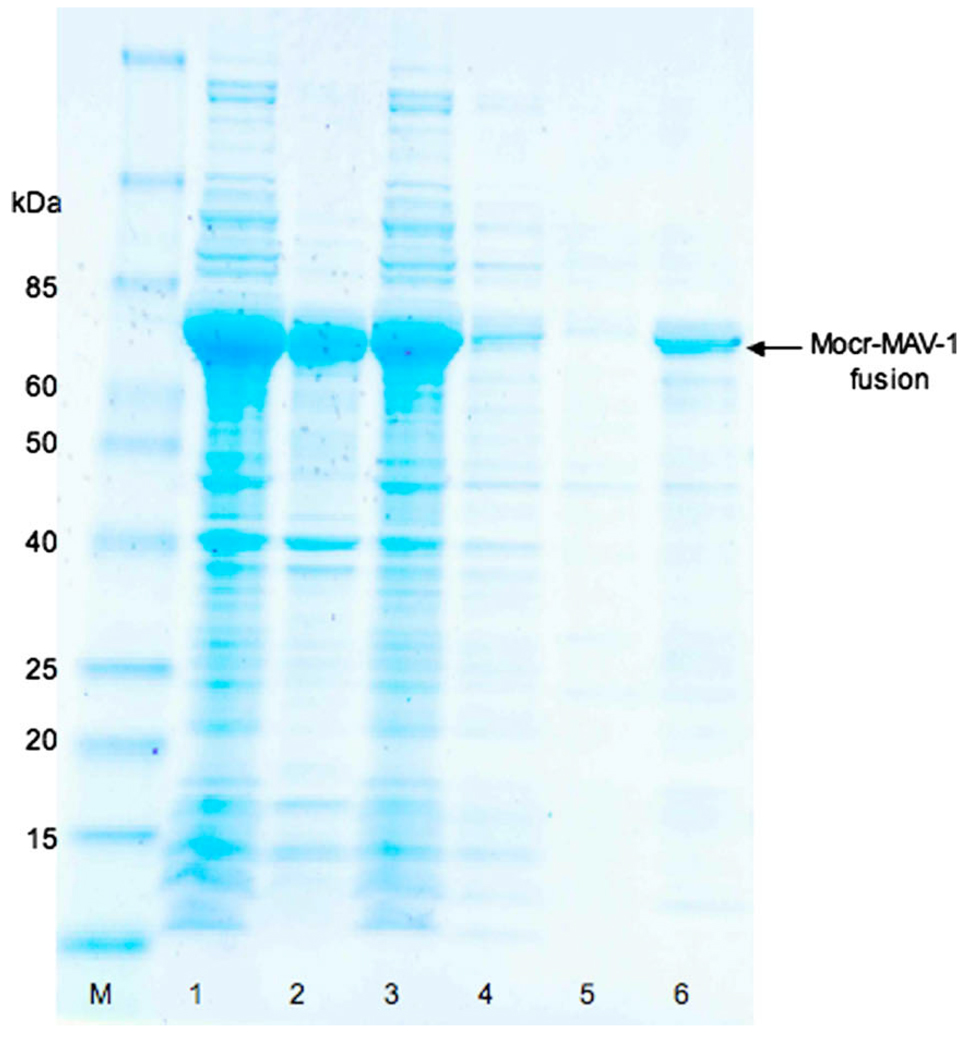
We then examined the effect of a passenger protein on the DEAE-cellulose purification profile of Mocr (Fig. 7). For these experiments, we used fusions of Mocr with one of the MAV-1 penton protein constructs (pI = 7.3) used in Table 1. The fusion was produced as described above for Mocr alone. The soluble fraction was batch bound to DEAE-cellulose resin, poured into a column, washed with a buffer containing 200 mM NH4Cl and eluted with one volume of buffer containing 600 mM NH4Cl. The eluted fusion protein displays a high level of purity, greater than 80%. This shows that the Mocr protein may function as a purification handle similar to other affinity fusion tags.
Fig. 7.
SDS–PAGE analysis of DEAE-cellulose purification fractions for a Mocr-MAV-1 fusion protein. Molecular weight marker (M), whole cell extract (1), insoluble fraction (2), soluble fraction (3), flow-through (4), wash (5), elution with 600 mM NH4Cl (6). The gel was stained with Coomassie blue. Arrow indicates the position of the fusion protein.
Cleavage of the passenger protein from the Mocr fusion
Med15 is a component of the Saccharomyces cerevisiae Mediator complex that is involved in activation of transcription [21]. In an effort to define interaction domains within this protein 72 different constructs were made which were fused with both MBP and Mocr. We have observed instances in which removal of MBP after cleavage from the fusion has proven difficult using affinity chromatography but could be accomplished by size-exclusion chromatography. In cases where the passenger and fusion tag are of similar size, this option is not available. This was the case with this Med15 construct; cleaved MBP could not be efficiently removed from the Med15 portion of the fusion (data not shown). In pMCSG9 [11], MBP is fused to a His6 tag allowing the MBP to be removed from the passenger protein after cleavage of a fusion protein by passing the preparation over a nickel column. A large portion of the Med15 was retained along with the MBP when this was done using this fusion construct, dramatically reducing the yield. The similarity of the molecular weights of the MBP and this Med15 construct did not allow for efficient separation using size exclusion chromatography.
This Med15 protein-Mocr fusion was tested for cleavage by the tobacco etch virus (TEV) protease and purification of the Med15 protein construct away from the Mocr portion of the fusion (Fig. 8). Cleavage of the Med15-Mocr fusion by TEV protease was complete; no intact fusion protein was visible after the reaction (Fig. 8A). The Med15 protein construct can then be purified away from the Mocr portion of the fusion by size-exclusion chromatography (Fig. 8B).
The use of Mocr with a passenger protein of this size allows for greater flexibility in this step of the purification protocol than a corresponding MBP fusion would allow.
Comparison of Mocr with another small fusion protein
Small fusion tags are desirable for many applications in which the fusion is left intact. We compared Mocr with GB1, another small fusion partner [22,23], in their ability to produce soluble variants of S11, a small (129 amino acids) ribosomal protein from E. coli. The E. coli protein was fused with His6 (pMCSG7), GB1 (cloned into pMCSG7 identically to mocr) or Mocr. The Mocr fusion solubilized almost all of the variants and yielded consistently higher levels of protein after nickel purification (Table 2).
Table 2.
Comparison of soluble protein yields between small fusion tags
| Constructa | Eluate conc. ng/µL (Total nmol)b | ||
|---|---|---|---|
| His | GB1 | Mocr | |
| T1-A | 210 (750) | 395 (1000) | 321 (584) |
| T1-B | 43 (160) | ndc | 596 (1084) |
| T1-C | 4 (13) | 304 (714) | 248 (428) |
| T1-D | 17 (64) | 39 (98) | 231 (420) |
| T1-E | 20 (74) | nd | nd |
| T1-F | 5 (17) | nd | 36 (62) |
| T3-A | 124 (462) | 485 (1227) | 776 (1412) |
| T3-B | 25 (93) | 491(1242) | 398 (720) |
| T3-D | 25 (93) | 77 (195) | 315 (573) |
| T3-E | nd | nd | 78 (142) |
| T4-A | 188 (700) | 381 (964) | 375 (680) |
| T4-B | 32 (119) | 254 (642) | 485 (882) |
| T4-D | 15 (56) | 31 (78) | 255 (464) |
| T4-E | 158 (588) | 36 (91) | 245 (446) |
T1, T3 and T4 represent three different mutant versions for the S11 ribosomal protein gene from E. coli. T1 contains a single amino acid substitution from the native sequence, T3 contains two and T4 contains eight. The same start and stop sites were used for each template but the C and F combinations were not made for T3 and T4.
96-well expression cultures followed by chemical lysis and metal-affinity chromatography were run as described in Materials and methods. The samples were then run on the Labchip 90 protein chip. The protein quantitation from the Labchip data was downloaded into an Excel file and is shown here. The total nmols for each sample was then calculated using these concentrations, the total volume and the molecular weight of each fusion protein.
Not detected.
Discussion
Limited solubility is one of the challenges in production of recombinant protein. Protein fusion partners are commonly used to solve solubility problems, and MBP is one of the most successful soubilizing partners for bacterially expressed proteins [7,8]. Although an excellent solubilizer, MBP is large (42 kDa) and may interfere with activity assays of the passenger protein. Some passengers interfere with the ability of MBP to bind amylose resin, and some proteins fall out of solution when MBP is removed from the fusion. Occasionally it is difficult to purify the passenger protein away from MBP after the fusion is cleaved. We set out to develop a novel fusion tag with solubilizing activity similar to MBP. We were particularly interested in developing a tag that was smaller than MBP and thus would interfere less with downstream applications and may not need to be removed from the purified fusion protein.
We have shown here that a monomeric form of the Ocr protein from bacteriophage T7 displays solubilizing activity when used as a fusion tag. The vector containing this fusion tag is compatible with high-throughput cloning and expression processes.
The best fusion tags also function as affinity tags, that is, they may be used for purification of the passenger protein. The most commonly used affinity tag is the His6 tag. This tag binds to immobilized transition metals and this is commonly used to purify the protein of interest from the cell lysate. MBP is also an affinity tag. It can be bound to amylose resin. The efficiency of this binding interaction may be reduced by the addition of a passenger protein to the MBP and so a His6 tag is frequently added to MBP fusions. In many cases this allows for high levels of purity to be attained because two affinity steps are available. Mocr protein may also be used as a purification handle with performance similar to affinity tags such as His6. The tag may be cleaved from its passenger protein using TEV protease and purified from the target protein by metal affinity, DEAE-cellulose or size exclusion chromatography. Uncleaved fusion protein may be fully active and in some cases may be compatible with assays of target protein function. Formation of dimers and receptor binding by the P domain of the MNV capsid protein and its ability to compete with native MNV in infectivity assays was not hindered by the presence of the Mocr fusion tag (Christiane Wobus, pers. com.). Mocr is thus a useful addition to available fusion tag strategies.
Numerous fusion proteins have been described but none is universally successful in solubilizing passenger proteins. Comparative studies have found wide variations in the performance of commonly used fusion tags [7,8]. The properties that give a fusion tag solubilizing activity are not understood. In a recent report of two new fusion tags, E. coli protein Skp was thought to function through chaperone activity, but the mechanism for bacteriophage T7 protein kinase was not clear [24]. Data for mutant forms of MBP are also consistent with a chaperone-like mechanism for solubilizing passenger proteins [25]. Another recent report attributed solubilizing activity to the acidity or charge of the fusion tag, E. coli protein Msb [10], and there have been previous suggestions that this may be an important property of solubilizing fusion tags [9]. Wilkinson and Harrison developed a statistical model for predicting protein solubility in bacteria [26]. A simplified version of this model, based on approximate charge average and turn-forming residue content, was developed as a predictor of suitable fusion proteins and used to identify the N-utilization substance A (NusA) protein [27]. NusA was predicted to have a 95% probability of solubility and then demonstrated to have solubilizing activity as a fusion tag. This model predicts Mocr to have a 97% probability of solubility and it also displays solubilizing activity.
Although the mechanism of solubility enhancement by the Mocr protein is not known, its effectiveness suggests that acidity and/or charge may be an important factor. Mocr has an acidic pI and high charge, in common with other documented solubilizing fusion tags. For example, MBP, NusA, Msb and the bacteriophage T7 protein kinase also have acidic pIs. Although acidity appears to be an important parameter in solubility it is not the only factor. The calculated pIs for our test proteins are: caveolin, 5.70; acyl transferase, 8.35; MAV-1 fiber knob, 5.05; MAV-1 penton, 7.35; MNV major capsid protein, 4.70. Three of the five proteins have acidic pIs and yet they display poor solubility when expressed in E. coli. It is unlikely that reducing the pI of the target protein by the addition of the acidic fusion tag is sufficient by itself to increase its solubility.
One mechanism of Mocr action may be its ability to reduce the aggregation of the passenger protein through Mocr’s high charge. The addition of charged acidic tails to aggregation-prone proteins has been shown to have this effect [28]. Charge repulsion between the acidic tails is thought to disrupt aggregation of the passenger protein. Such a charge-repulsion effect may be partially responsible for the solubility observed using the Mocr protein fusion. The Mocr protein has the additional advantage of being a stable, well-structured protein, unlike the acidic tails, which are most likely unstructured.
Since it is not possible to predict a priori which tag may be useful with a given protein, many groups have adopted a multi-parallel strategy containing several fusion tags, which is made possible by the use of high-throughput formats. The Mocr fusion is now one of the tags we routinely use when identifying conditions for soluble expression of various constructs of target proteins.
Although the Mocr fusion tag may be efficiently removed from the passenger protein, for some applications it may be preferable to leave the fusion intact. One such application is NMR analysis. As cloned in pMCSG7, our Mocr fusion protein adds 16.7 kDa to the passenger protein or peptide. MBP, as it is cloned into the pMCSG9 vector, adds 43.6 kDa to the passenger protein or peptide. In a uniformly labeled protein for use in NMR experiments, the additional mass of the MBP would greatly complicate the analysis and would require removal. A need to remove the fusion tag introduces a variety of complications and led to the development of smaller fusions partners such as GB1 [22,23] that do not require removal before spectroscopic analysis. Although larger than GB1, Mocr’s size is consistent with the Domain I of E. coli initiation factor 2 (IF2, 17.4 kDa) that has also been proposed as a solubility fusion tag for use in NMR applications [29].
Given the small size and compactness of the Mocr structure, it may be possible to crystallize the passenger protein while still fused to the Mocr protein. This has not been reported with larger fusion tags, such as GST and MBP, unless the passenger protein was very small, less than 115 amino acids [30]. Crystallization trials with Mocr fusions are underway to investigate this possibility.
In the crystal structure of Ocr protein, only residues 5–110 were ordered, of a total of 116 amino acids. When we cloned the synthetic gene into the pMCSG backbone we provided no additional linker between the Mocr and the TEV cleavage site because the disorder of the last six amino acids of Mocr should allow for protease access to the cleavage site. In a number of different Mocr fusions, we observed complete cleavage of the fusion with TEV protease. The four residues that are disorded at the Ocr amino terminus should also provide an unstructured linker region to facilitate accessibility for TEV or other protease cleavage in a carboxyl fusion of Mocr.
Inclusion body formation in E. coli displays significant similarity to amyloid plaque formation [31]. Since aggregation of target protein may be driven by surface hydrophobicity as in plaque formation [31], a strategy to reduce this effect could be the introduction of less polar solvents, such as ethanol, into lysis buffers. This would be analogous to work with amyloid-forming peptides for which addition of organic solvents such as ethanol (10%) and DMSO (5%) has the effect of disaggregating protein and stabilizing monomers [32]. Organic solvents may reduce aggregation but they may have a negative effect on overall protein product solubility. One of the features that initially attracted us to Ocr was the observation of its solubility in 95% ethanol [14]. Thus, Mocr should be soluble in a less polar solvent at the relatively low concentrations at which disaggregation has been observed for amyloid forming peptides, possibly stabilizing a passenger protein as a monomer in the soluble fraction of the lysate. We have some preliminary data using up to 10% ethanol suggesting this is a viable strategy and an area for further investigation.
Acknowledgments
The HTP lab has been supported in part by a grant from the Office of the Vice-President of Research at the University of Michigan. C.Y.M. is supported by NIH Grant GM65330 to Dr. Anna Mapp. We are grateful to Dr. John Louis of NIDDK at NIH for providing GEV2 as a source for GB1. We are also grateful to our collaborators, Dr. Alan Saltiel, Dr. David Sherman, Dr. Kathy Spindler, Dr. Christiane Wobus, Dr. Jeanne Stuckey, and Dr. Anna Mapp, all of the University of Michigan, whose projects contributed to the work described here.
Footnotes
Abbreviations used: Mocr, monomeric Ocr; MBP, maltose-binding protein; LIC, ligation-independent cloning; TEV, tobacco etch virus; GST, glutathione-S-transferase; MAV-1, mouse adenovirus type 1; MNV, mouse norovirus.
References
- 1.Hedren M, Ballagi A, Mortsell L, Rajkai G, Stenmark P, Sturesson C, Nordlund P. GRETA, a new multifermentor system for structural genomics and process optimization. Acta Cryst. 2006;62:1227–1231. doi: 10.1107/S090744490603441X. [DOI] [PubMed] [Google Scholar]
- 2.Goulding CW, Perry LJ. Protein production in Escherichia coli for structural studies by X-ray crystallography. J. Struct. Biol. 2003;142:133–143. doi: 10.1016/s1047-8477(03)00044-3. [DOI] [PubMed] [Google Scholar]
- 3.Cabrita LD, Dai W, Bottomley SP. A family of E. coli expression vectors for laboratory scale and high-throughput soluble protein production. BMC Biotechnol. 2006;6 doi: 10.1186/1472-6750-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2006;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 6.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammarstrom M, Hellegren N, Van Den Berg S, Berglund H, Hard T. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 2002;11:313–321. doi: 10.1110/ps.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox JD, Routzhn KM, Butcher MH, Waugh DS. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 2003;537:53–57. doi: 10.1016/s0014-5793(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 10.Su Y, Zou Z, Fend S, Zhou P, Cao L. The acidity of protein fusion partners predominately determines the efficacy to improve the solubility of the target proteins expressed in Escherichia coli. J. Biotechnol. 2007;129:373–382. doi: 10.1016/j.jbiotec.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Donnelly MI, Zhou M, Sanville Millard C, Calncy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryor KD, Leiting B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding protein double-affinity fusion system. Protein Expr. Purif. 1997;10:309–319. doi: 10.1006/prep.1997.0759. [DOI] [PubMed] [Google Scholar]
- 13.Dunn JJ, Elzinga M, Mark K-K, Studier FW. Amino acid sequence of the gene 0.3 protein of bacteriophage T7 and nucleotide sequence of its mRNA. J. Biol. Chem. 1981;256:2579–2585. [PubMed] [Google Scholar]
- 14.Mark K-K, Studier FW. Purification of the gene 0.3 protein of bacteriophage T7, an inhibitor of the DNA restriction system of Escherichia coli. J. Biol. Chem. 1981;256:2573–2578. [PubMed] [Google Scholar]
- 15.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. Structure of Ocr from Bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 16.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a Tobacco Etch Virus protease cleavage site. Protein Expr. Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 17.Kruger DH, Schroder C, Hansen S, Rosenthal HA. Active protection by Bacteriophages T3 and T7 against E. coli B- and K-specific restriction of their DNA. Mol. Gen. Genet. 1977;153:99–106. doi: 10.1007/BF01036001. [DOI] [PubMed] [Google Scholar]
- 18.Atanasiu C, Byron O, McMiken H, Surrock SS, Dryden DTF. Characterization of the structure of ocr, the gene 0.3 protein of bacteriophage T7. Nucleic Acids Res. 2001;29:3059–3068. doi: 10.1093/nar/29.14.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez I, Ying Y, Albanesi J, Anderson RG. Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. USA. 2002;99:11193–11198. doi: 10.1073/pnas.172196599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2006;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Huth JF, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koenig BW, Rogowski M, Louis JM. A rapid method to attain isotope labeled small soluble peptides for NMR. J. Biomol. NMR. 2003;26:193–202. doi: 10.1023/a:1023887412387. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee DK, Esposito D. Enhanced soluble protein expression using two new fusion tags. Protein Expr. Purif. 2006;46:122–129. doi: 10.1016/j.pep.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 25.Nallamsetty S, Waugh DS. Mutations that alter the equilibrium between open and closed conformations of Escherichia coli maltose-binding protein impede its ability to enhance the solubility of passenger proteins. Biochem. Biophys. Res. Commun. 2007;364:639–644. doi: 10.1016/j.bbrc.2007.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Bio/Technology. 1991;9:443–448. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- 27.Davis GD, Elisee C, Newham DM, Harrison RG. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 1999;65:382–388. [PubMed] [Google Scholar]
- 28.Zhang Y-B, Howitt J, McCorkle S, Lawrence P, Springer K, Freimuth P. Protein aggregation during overexpression limited by peptide extensions with large net negative charge. Protein Expr. Purif. 2004;36:207–216. doi: 10.1016/j.pep.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Sorenson HP, Sperling-Peterson HU, Mortensen KK. A favorable solubility partner for the recombinant expression of streptavidin. Protein Expr. Purif. 2003;32:252–259. doi: 10.1016/j.pep.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12:1313–1322. doi: 10.1110/ps.0243403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrio M, Gonzalez-Montalban N, Vera A, Villaverde A, Ventura S. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 2005;347:1025–1037. doi: 10.1016/j.jmb.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 32.Sabate R, Estelrich J. Disaggregating effects of ethanol at low concentration on beta-poly-l-lysines. Int. J. Biol. Macromol. 2003;32:10–16. doi: 10.1016/s0141-8130(03)00019-9. [DOI] [PubMed] [Google Scholar]