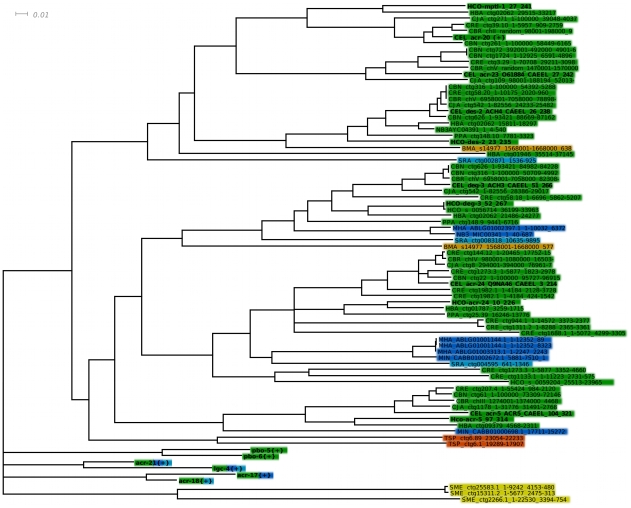
Figure 2. Detailed view of the DEG-3 sub-family.
Detailed view of the DEG-3 sub-family from the LBD region NJ tree, with branches below 50% support after 1000 bootstrap iterations joined. A few related subfamilies are shown as collapsed branches. CEL, Caenorhabditis elegans, CBN, C. brenneri, CBR, C. briggsae, CRE, C. remanei, CJA, C. japonica, PPA, Pristionchus pacificus, HCO, Haemonchus contortus, HBA, Heterorhabditis bacteriophora, MHA, Meloidogyne hapla, NM3MIC/MIN, M. incognitia, NB3AYC Ancylostoma ceylanicum, SRA, Strongyloides ratti, BMA, Brugia malayi, TSP, Trichinella spiralis, SME, Schmidtea mediterranea. Pristionchus pacificus lacks a close MPTL-1 homolog and was predicted to be less sensitive to AAD-1566 than species such as C. japonica, H. contortus and H. bacteriophora.