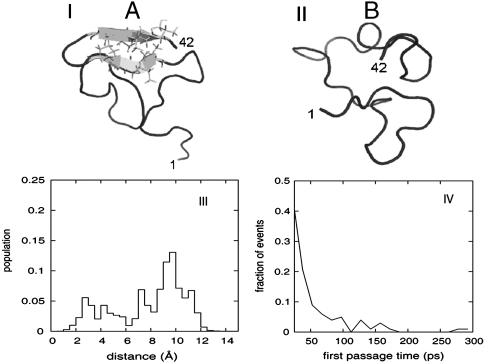
Fig. 1.
I and II) Representative conformations of the two ensemble populations of Aβ (1–42) used to model the locally structured “A” and disordered “B” states, as extracted from the replica exchange molecular dynamics (REMD) trajectories. Atoms of residues that participate in the C-terminal β-hairpin are explicitly shown. The first and last residues of each conformation are numbered for clarity. Graphics were generated by mean of the visualization suite pymol (www.pymol.org). (III) Distribution of atomistic (Cα) rmsd from the representative conformation of the B (locally ordered) state, as calculated for all REMD conformations sampled at low temperature. All conformations were sampled over a range of temperatures from 276 K to 305 K. A bimodal distribution with a gap at 6 Å indicates the existence of two populations with distinct structural characteristics. (IV) Distribution of transition times (first passage time) between the A (locally ordered) and B (disordered) states, as observed in the low-temperature segments of the REMD trajectories. All events happen over a range of temperatures from 276 K to 305 K. The majority (> 80%) of events take place in the sub-100 ps time scale, which makes our dataset suitable for analysis with the 2DIR simulation method.