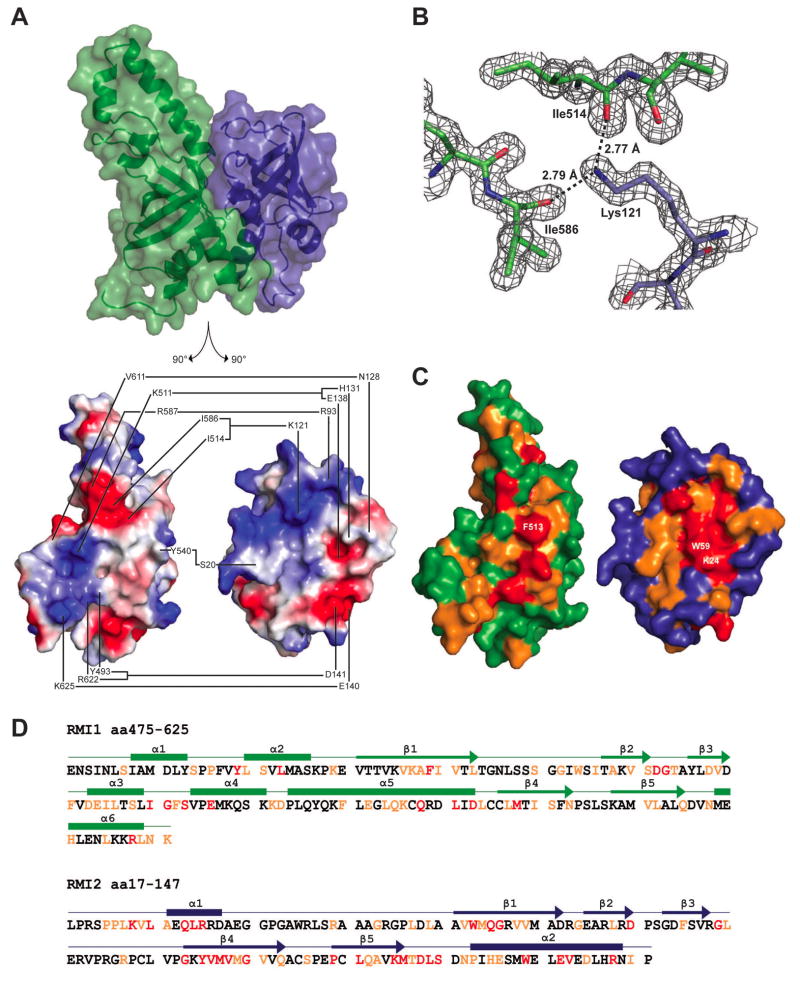
Figure 2. RMI core complex interface.
(A) Top: structure of the RMI core complex showing semi-transparent surface and ribbon representations in the same orientation and color scheme as in 1B. Bottom: split view of the RMI core complex in which both proteins are separated and rotated 90° to show interacting surfaces. The interface between RMI1 and RMI2 is shown with connected lines indicating residues forming hydrogen bonds or ionic interactions. The interface is depicted using surface electrostatics, showing electropositive (blue) and electronegative (red) surface potential. (B) 2Fo − Fc electron density map (1.5 σ) showing interaction of RMI1 Ile514 and Ile586 with RMI2 Lys121. (C) The interface between RMI1 and RMI2 is shown in the same orientation as 2A, bottom. RMI1 and RMI2 are colored to indicate residues that are invariant (red), highly conserved (orange), or poorly conserved (green for RMI1 and blue for RMI2) among identified RMI1 and RMI2 proteins from human, bovine, mouse, zebrafish, and arabidopsis sequences. Three of the invariant residues that form a hydrophobic interaction network are labeled: RMI1 Phe513, RMI2 Lys24, and RMI2 Trp59. (D) The sequence for the RMI core complex is shown, with secondary structural features labeled above as boxes (α-helices) and arrows (β-strands). Red and orange lettering indicates invariant and highly conserved residues, respectively. See also Figure S2.