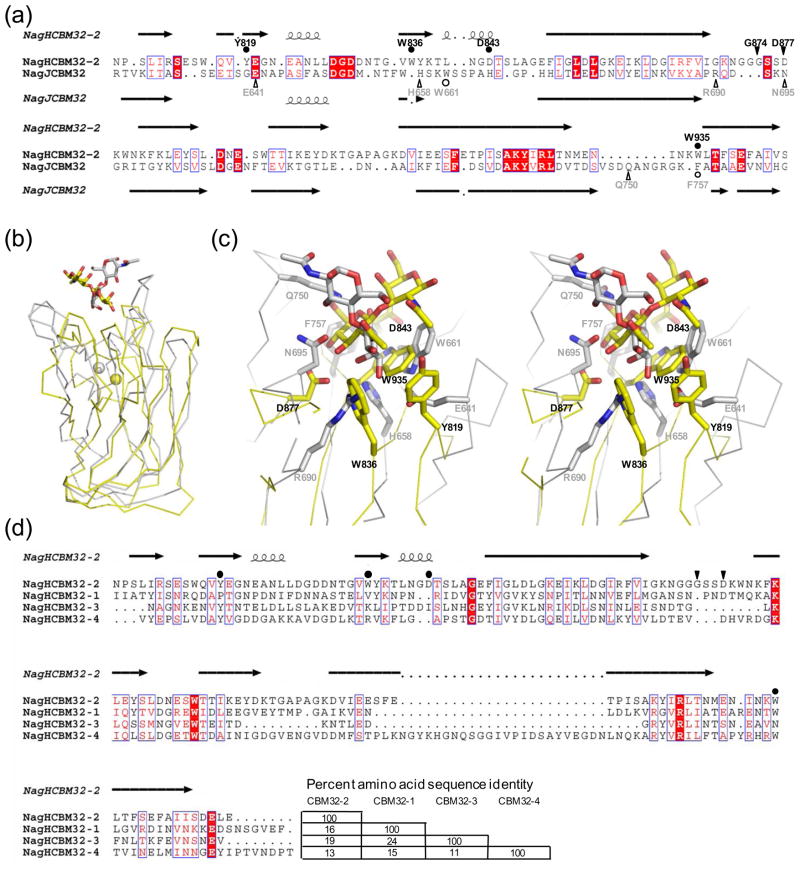
Figure 4.
Comparison of NagHCBM32-2 with other family 32 carbohydrate-binding modules. (a) Amino acid sequence alignment of NagHCBM32-2 with the CBM, NagJCBM32, from C. perfringens NagJ based on a structural overlay produced by secondary structure matching 28. Secondary structures for NagHCBM32-2 and NagJCBM32 are shown above and below the alignment, respectively. Residues in NagHCBM32-2 involved in hydrogen bonds with the carbohydrate are indicated by filled triangles and labeled. Residues making primarily van der Waals interactions are indicated with filled circles. Residues performing these functions in NagJCBM32 are indicated below the alignment in open symbols. (b) Structural overlay of NagHCBM32-2 (yellow) and NagJCBM32 (grey). Bound sugars are shown in stick representation and metal ions as spheres. (c) The overlapped active sites of NagHCBM32-2 (yellow) and NagJCBM32 (grey) shown in divergent stereo. Bound ligands and residues that interact with these are shown in stick representation. Residues in NagHCBM32-2 are labeled in black and those in NagJCBM32 labeled in grey. (d) Amino acid alignment of NagHCBM32-2 with the other putative CBM32s in NagH. This alignment was produced using ClustalW 53. The secondary structure and functional residues of NagHCBM32-2 are indicated as in panel (a). Amino acid sequence identities of the modules are shown at the end of the alignment. The alignments in panels (a) and (d) were displayed using ESPript 54.