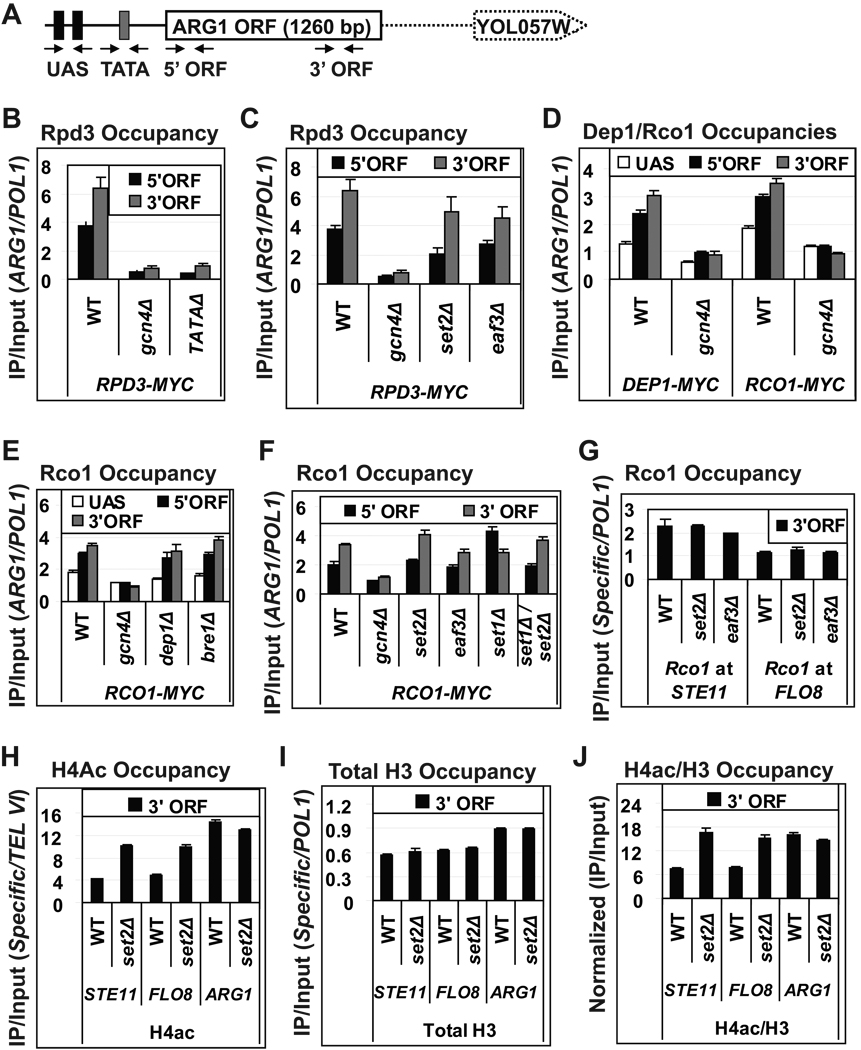
Fig. 1. Co-transcriptional recruitment of Rpd3C(S) is independent of H3-K36 methylation and Eaf3.
(A) Location of primers for ChIP analysis of ARG1. (B–F) Occupancies of Rpd3-myc, Dep1-myc and Rco1-myc at ARG1 were measured by ChIP in the indicated strains cultured in SC medium and treated with sulfometuron methyl (SM) for 30 min to induce Gcn4. Cross-linked chromatin was immunoprecipitated with anti-myc antibodies and DNA from immunoprecipitated (IP) and input samples was subjected to PCR in the presence of [33P]-dATP to amplify radiolabeled fragments of ARG1 or the POL1 ORF, analyzed as a control, which were resolved by PAGE and quantified by phosphorimaging. Ratios of ARG1 to POL1 signals in IP samples were normalized to the ratios for Input samples to yield occupancy values. (G) Occupancies of Rco1-myc at the 3’ ORF regions of STE11 and FLO8 measured by ChIP in the indicated strains. (H–I) Occupancies of acetylated H4 (H4Ac) and total H3 at the 3’ ORFs of STE11, FLO8 and ARG1 were measured by ChIP in cells grown in YEPD, using primers for these genes and non-transcribed sequences at the right-arm telomere of VI (TEL VI) (H) or POL1 (I). (J) H4Ac occupancies from (H) normalized for total H3 (I). All error bars represent the standard error of mean (SEM). For Rpb3 occupancies in TATAΔ and set2Δ see Fig. S1A.