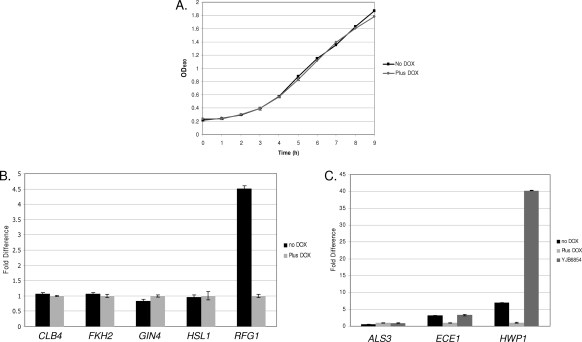
Fig. 8.
(A) Growth curves of the tet-RFG1 strain. The tet-RFG1 strain was grown overnight in the presence of doxycycline, and cells were washed before being diluted 1:30 in fresh YPD or YPD plus DOX. Duplicate cultures were grown with shaking at 30°C, samples were removed at the times indicated, and the OD600 was measured. The strain grew at identical rates regardless of the presence or absence of doxycycline. (B) Quantification of mitotic regulatory gene transcription in the presence of elevated RFG1. The tet-RFG1 strain was grown overnight in the presence of doxycycline, and cells were washed before being diluted 1:20 into fresh YPD and grown for 6 h at 30°C in the presence or absence of doxycycline. At this point the culture grown in the presence of doxycycline consisted entirely of yeast cells, while the culture grown in the absence of doxycycline was approximately 80% pseudohyphal. RNA was isolated from cells harvested from these cultures, and transcript levels were quantified through real-time PCR analysis. Levels were normalized by ACT1 and are expressed relative to the control tet-RFG1 plus doxycycline culture (therefore, its results are 1.0). While RFG1 levels are elevated approximately 4.5-fold in the tet-RFG1 strain grown in the absence of doxycycline, there are only minor differences in transcript levels of the other genes examined. Error bars represent the standard deviations of the threshold cycle (ΔCT) values. (C) Quantification of filament-specific gene transcription in the context of elevated RFG1 expression. The tet-RFG1 and the clb4Δ null (YJB6854) strains were grown, and transcript levels were analyzed as described for panel B. Transcription of these genes followed the same pattern in both the tet-RFG1 strain grown in the absence of doxycycline and the clb4Δ null strain.