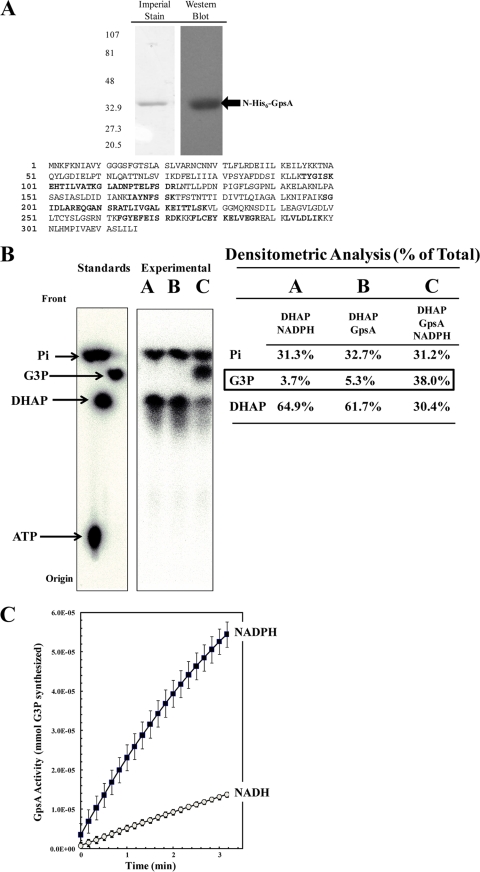
FIG. 1.
Recombinant N-His6-GpsA protein is an active G3P dehydrogenase in vitro. R. prowazekii gpsA was cloned into pET15b, heterologously expressed in E. coli BL21(DE3), and purified with N-His6-GpsA. (A) Purified protein was resolved on a 4 to 15% Tris-HCl SDS-polyacrylamide gel and visualized by Imperial protein staining and Western blot analysis using anti-His6 monoclonal antibody (sizes [in kDa] are indicated on the left). The mass spectrometry analysis coverage map shows matched peptides in bold font (30% coverage). (B) N-His6-GpsA-catalyzed conversion of [32P]DHAP to [32P]G3P was verified by paper chromatography. [32P]ATP, [32P]DHAP, [32P]G3P, and [32P]orthophosphate were used as standards. The reaction mixtures were incubated for 60 min at room temperature prior to chromatographic analysis. Lane A, a negative-control reaction mixture containing 6 μM [32P]DHAP and 100 μM NADPH only; lane B, a negative-control reaction mixture containing [32P]DHAP and 2.5 μg ml−1 N-His6-GpsA protein only; lane C, the N-His6-GpsA-catalyzed conversion of [32P]DHAP to [32P]G3P absolutely required the presence of NADPH. The density of each compound on the chromatograph is expressed as a percentage of the total lane density (summarized in tabular form to the right of each spot on the chromatograph). (C) N-His6-GpsA-catalyzed conversion of DHAP to G3P was measured spectrophotometrically at OD340 by following the oxidation of NADPH or NADH over time. Error bars represent standard deviations. The reaction mixtures contained 1.0 μg ml−1 N-His6-GpsA, 500 μM DHAP, and 100 μM NAD(P)H in modified buffer A; and the reactions were carried out at room temperature.