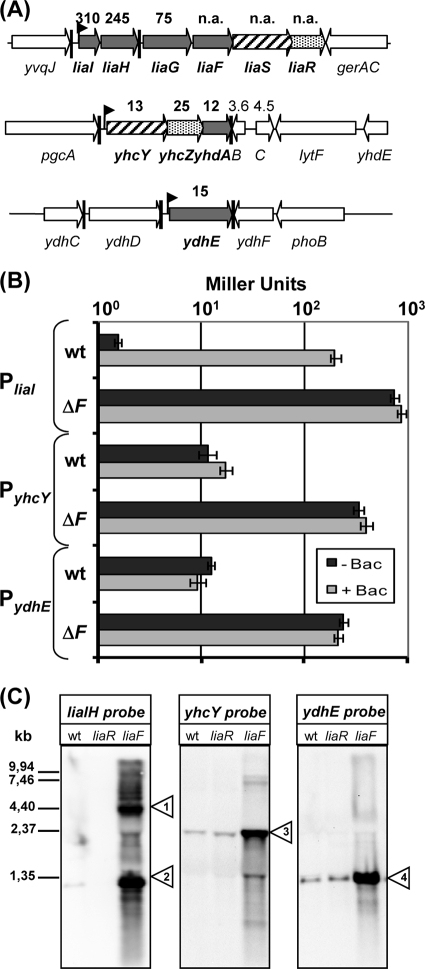
FIG. 1.
Identification of LiaR-dependent target genes. (A) Genomic organization of potential LiaR target genes as identified by DNA microarray analysis. Expression of the liaIHGFSR operon, yhcYZ-ydhA operon, and the ydhE gene are strongly induced in a Lia ON mutant relative to their expression in the wild-type strain (see fold-change values above the arrows; n.a., values derived for these genes are an artifact due to replacing the liaF gene with the kanamycin resistance cassette). (B) Measurement of the β-galactosidase activity of the PliaI, PyhcY, and PydhE promoters in the wild type (wt) and the corresponding liaF::kan background. Cultures of strains TMB016 (PliaI-lacZ), TMB018 (PliaI-lacZ liaF::kan), TMB071 (PyhcY-lacZ), TMB095 (PyhcY-lacZ liaF::kan), TMB222 (PydhE-lacZ), and TMB224 (PydhE-lacZ liaF::kan) were grown in LB medium to mid-log phase and then split. One half was induced by the addition of bacitracin (+ Bac; 50 μg ml−1 final concentration), and the other half served as an uninduced control (− Bac). Cells were harvested after 30 min of incubation and assayed as described previously (46). β-Galactosidase activity is expressed in Miller units (47). Error bars show standard deviations. (C) Northern blot analysis. Expression of liaIHGFSR, yhcYZ-yhdA, and ydhE was measured using 5 μg of total RNA from each strain sample (wild type, TMB002 [liaF::kan], and HB0933 [liaR::kan]) separated on a 1.2% formaldehyde gel. RNA was transferred to a nylon membrane and hybridized with labeled gene-specific RNA probes (see Materials and Methods for details). Small triangles indicate the major transcript(s) for each gene; their sizes correspond to transcripts liaIHG-kan-liaSR (1), liaIH (2), yhcYZ-ydhA (3), and ydhE (4).