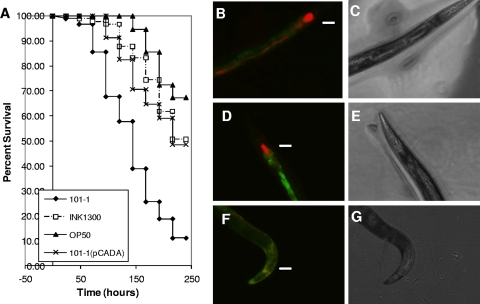
FIG. 6.
(A) C. elegans survival rates of wild-type EAEC outbreak strain 101-1, 101-1 complemented with pCADA, and INK1300, the derivative of 101-1 with the cad operon integrated into the chromosome. Strain OP50 was used as a negative control. Both strain INK1300 and 101-1 bearing pCADA were significantly attenuated compared to the wild-type 101-1 strain (P = 0.001). The observed differences between strains fed on INK1300 and 101-1(pCADA) did not show statistical significance. (B to G) Visualization of bacterial strains OP50, 101-1, and INK1300 expressing dsRed from pHKT4 inside infected C. elegans. (B) Fluorescence of bacteria within a worm that has used 101-1(pHTK4) as a food source. Note intense colonization of the pharynx and intact bacteria present inside the worm anterior to the grinder (marked with an arrow in panels B, D, and F). Autofluorescence of the lumen is visualized in green. (D) Fluorescence of a worm that has used INK1300(pHTK4) as a food source. As with 101-1, the pharynx is colonized but there are fewer bacteria posterior to the grinder. (F) Fluorescence of a worm that has used OP50 as a food source, showing very little pharyngeal colonization and no detectable red fluorescence distal to the grinder. (C, E, and G) Light field views of worms in panels B, D, and F, respectively. Photomicrographs were taken 3 days postseeding with a fluorescence microscope at ×100 magnification.