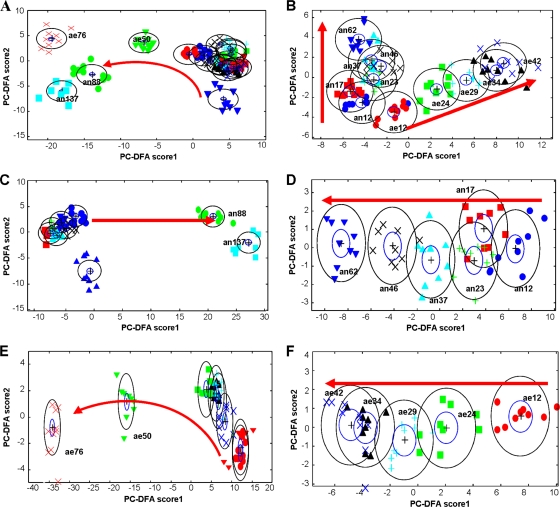
FIG. 3.
PC-DFA plots for FT-IR spectra. (A) Combined aerobic and anaerobic samples; (B) early-exponential- and stationary-phase samples grown aerobically and anaerobically; (C) combined anaerobic samples; (D) early-exponential- and stationary-phase samples grown anaerobically; (E) combined aerobic samples; (F) early-exponential- and stationary-phase samples grown aerobically. The red arrows represent an increase in incubation time. The number after the label stands for the incubation time; labels beginning with “ae” are for aerobic samples, and those beginning with “an” are for anaerobic samples. For example, an12 represents cells grown in anaerobic medium for 12 h, and ae24 represents cells grown in aerobic medium for 24 h. Dark blue ▴, an0; dark blue •, an12; red ▪, an17; green +, an23; light blue ▴, an37; black ×, an46; dark blue ▾, an62; green •, an88; light blue ▪, an137; red ▾, ae0; red •, ae12; green ▪, ae24; blue +, ae29; black ▴, ae34; dark blue ×, ae42; green ▾, ae50; red ×, ae76. PCs 1 to 20 (accounting for 99.5% of the total explained variance) were used by the DFA algorithm with a priori knowledge of machine replicates (i.e., 1 class per sample point, giving 17 classes in total). The different symbols represent the different sample points. The circles represent the 95% χ2 confidence interval about the mean.