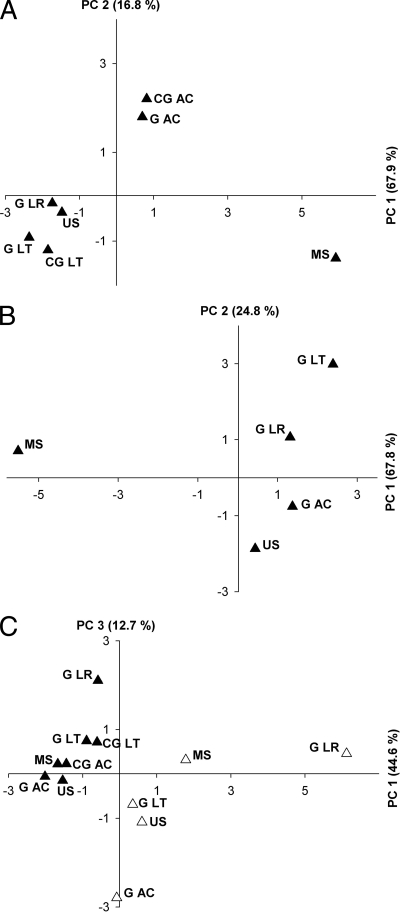
FIG. 3.
Principal-component analyses of TRFs. (A) narG TRFs (digestion was with BanI). A variance of 84.7% is covered by the x (67.9%) and y (16.8%) axes. (B) narG transcript TRFs (digestion was with MaeIII). A variance of 92.6% is covered by the x (67.8%) and y (24.8%) axes. (C) nosZ TRFs (filled symbols) and nosZ transcript TRFs (open symbols). Digestion was with HhaI. A variance of 57.3% is covered by the x (44.6%) and y (12.7%) axes. Abbreviations: G, gut; CG, crop/gizzard; AC, A. caliginosa; LT, L. terrestris; LR, L. rubellus; MS, mineral soil; US, uppermost soil.