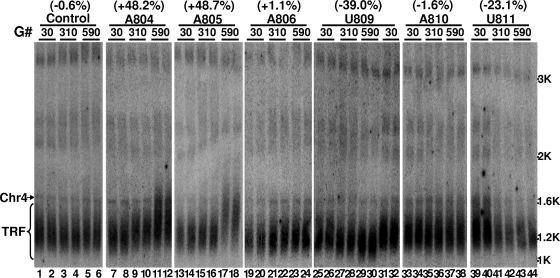
FIG. 5.
Southern analysis of chromosome end length in cells expressing various artificial guide RNAs. Cells (yCH-002) expressing a random gRNA (Control), gRNA-A804 (lanes 7 to 12), gRNA-A805 (lanes 13 to 18), gRNA-A806 (lanes 19 to 24), gRNA-U809 (lanes 25 to 32), gRNA-A810 (lanes 33 to 38), or gRNA-U811 (lanes 39 to 44) were harvested after being grown for the indicated number of generations (G#). DNA was recovered, digested with XhoI, and hybridized with two radiolabeled probes, a telomere-specific one and a chromosome IV-specific one (as an internal control). The fragment of chromosome IV (Chr4) and the telomeres (TRF) are indicated. For each strain, two independent experiments are presented in duplicate lanes. The distances between the signal peaks of Chr4 and TRF were measured using ImageQuant software (Molecular Dynamics). Shown in parentheses at the top of each strain are the relative changes in distance between two time points (30 and 590 generations) (difference in average distance between the two time points, divided by the average distance at 30 generations). “+” indicates telomere lengthening, and “−” indicates telomere shortening. Numbers at the right indicate sites of double-stranded DNA.