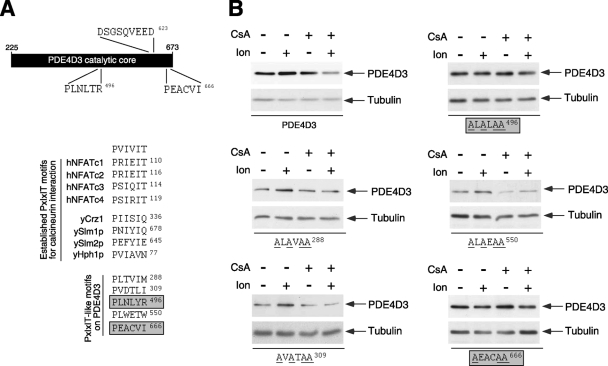
FIG. 4.
Mapping of functional PXIXIT-like calcineurin docking motifs in PDE4D. (A) Schematic illustration of the locations of the two functional PXIXIT-like motifs (PLNLYR496 and PEACVI666) in the catalytic core of PDE4D3. The location of the DSGSQVEED phosphodegron in PDE4D3 is also indicated. A sequence comparison of known calcineurin docking PXIXIT motifs found in human NFAT and yeast proteins is shown. Potential PXIXIT-like motifs in PDE4D3 and an optimized PXIXIT motif (PVIVIT) are also shown. Functional PXIXIT-like motifs in PDE4D3 are highlighted. (B) Wild-type and PXIXIT-mutated PDE4D3 were transiently transfected into COS7 cells. The expression levels of wild-type and PXIXIT-mutated PDE4D3 were determined in the presence and absence of Ion (2 μM) and/or CsA (5 μM). The expression of tubulin was used as a control. Replacements of critical amino acid residues with Ala in potential PXIXIT-like motifs of PDE4D3 are underlined. Functional PXIXIT-like motifs in PDE4D3 are highlighted.