FIG. 1.
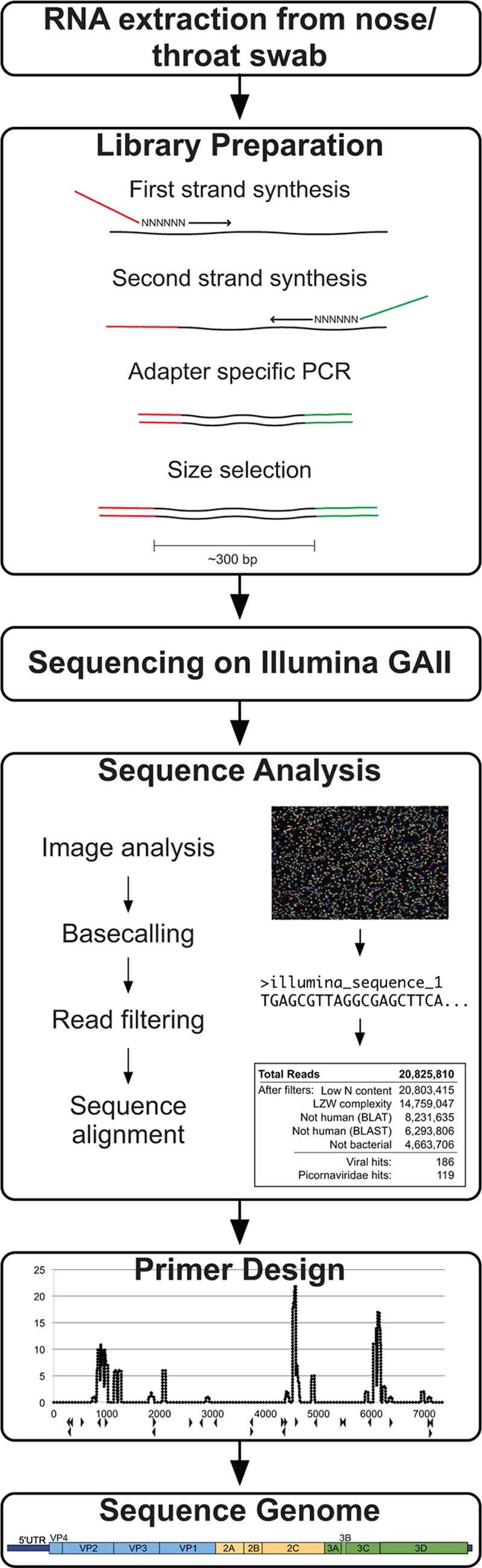
Viral recovery workflow. The method for viral genome recovery begins with RNA extraction from a nose/throat swab of clinical interest. A sequencing library is generated from the nucleic acids using reverse transcription and random-primed amplification to affix known sequencing adapter molecules necessary for the Illumina platform. The Illumina GAII sequencing is performed using the manufacturer's protocol. Sequence analysis includes image analysis and base-calling software, read filtering using quality metrics, and iterative sequence alignments. Reads with similarity to the viral sequence of interest are computationally assembled or used for primer design for subsequent specific amplification of remaining viral regions.
