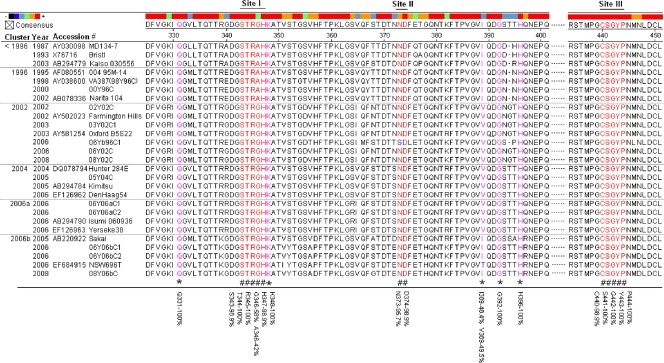
FIG. 4.
Sequence alignment of the HBGA-binding interfaces and the surrounding regions. The P domain sequences of 27 GII-4 viruses selected from different years were aligned using MegAlign. The numbers along the top indicate the position of the residues according to consensus sequence. Amino acids in red (noted by “#” at the bottom) indicate residues that are required for HBGA binding. Pink residues with “*” at the bottom indicate amino acids that affect the binding specificity. Blue letters indicate differences from the consensus sequence within the binding interfaces or surrounding regions. The percent similarity between all 93 strains is indicated at the bottom for key amino acid positions involved in HBGA binding.