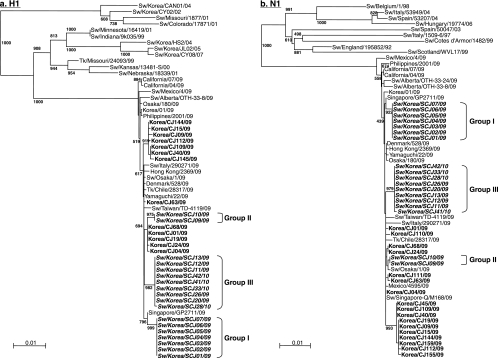
FIG. 1.
(a and b) Phylogenetic analysis of the HA and NA genes of pandemic (H1N1) 2009 influenza virus isolates from South Korea. Phylogenetic trees of the nucleotide sequences for the H1 (a) and N1 (b) genes of pandemic viruses isolated from pigs and humans in this study compared with nucleotide sequences from selected swine, human, and avian influenza virus strains available in GenBank are shown. The nucleotide sequences were aligned using Clustal_X (1, 38), and the phylograms were generated by the neighbor-joining method using the tree-drawing program NJ plot (31). The scale represents the number of substitutions per nucleotide. Branch labels record the stability of the branches over 1,000 bootstrap replicates. Only bootstrap values ≥400% are shown in each tree. Sw, swine; Tk, turkey.