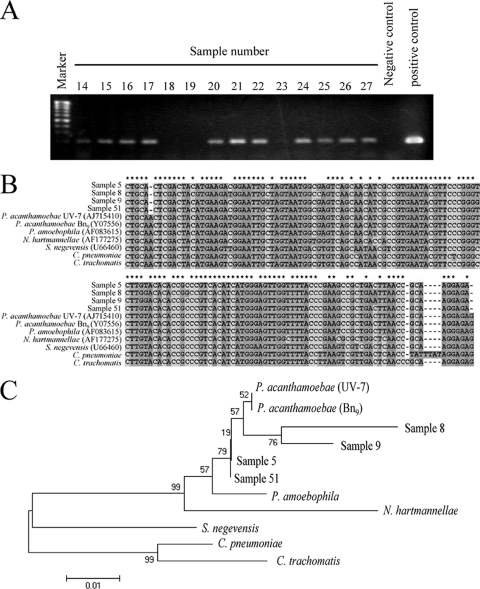
FIG. 1.
Detection and analysis of P. acanthamoebae DNA in smear samples obtained from a hospital environment. (A) Representative results of agarose gel electrophoresis of P. acanthamoebae PCR products amplified from smear samples. (B) Alignment analysis of four P. acanthamoebae amplicons (samples 5, 8, 9, and 51) with previously reported Parachlamydiaceae sequences was performed using Genetyx-Mac software (version 10.1). Asterisks represent conserved sequences in all P. acanthamoebae PCR amplicons. Dots and unmarked positions represent base mismatches and additional bases in the alignment, respectively. P. amoebophila, Protochlamydia amoebophila; N. hartmannellae, Neochlamydia hartmannellae. (C) Construction of a phylogenetic tree for P. acanthamoebae amplicons (samples 5, 8, 9, and 51) with previously reported Parachlamydiaceae sequences was performed using the neighbor-joining method in MEGA software (version 4). Strain names are in parentheses. The accession numbers assigned by DDBJ were as follows: sample 5, uncultured Parachlamydia sp. HUHP-5, AB565471; sample 8, HUHP-8, AB565472; sample 9, HUHP-9, AB565473; sample 51, HUHP-51, AB565474.