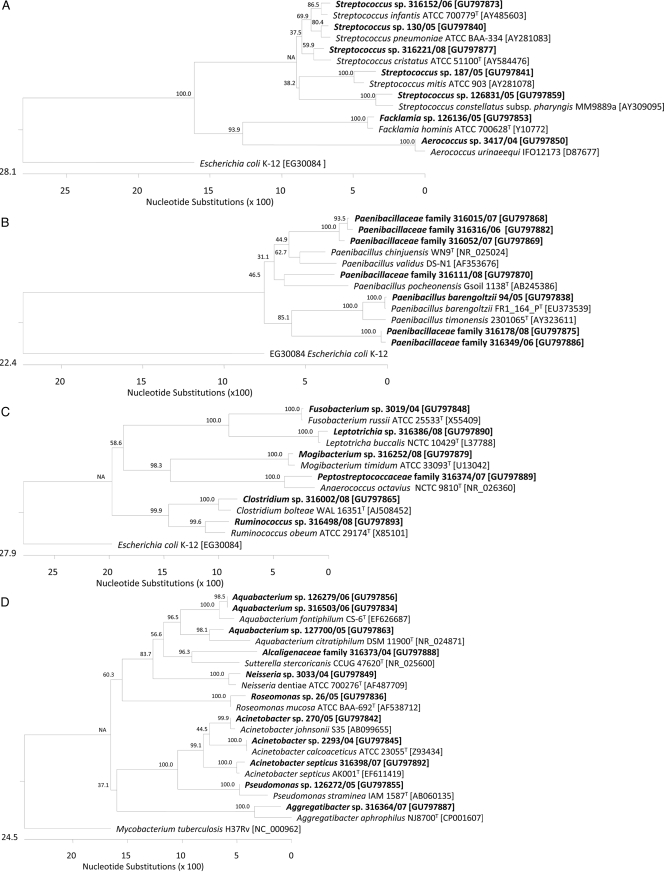
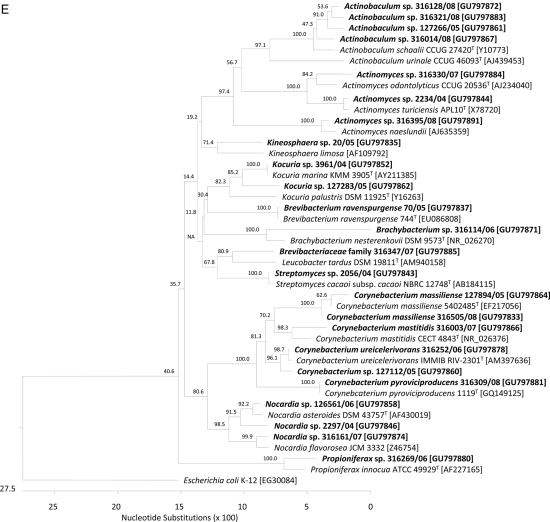
FIG. 1.
Phylogeny of 55 of 60 cultured isolates recovered from clinical specimens with homology of <99% to 16S rRNA gene sequences of members of published taxa. The dendrograms were calculated using CLUSTAL V alignment and a matrix of Jukes-Cantor distances determined by the neighbor-joining method using DNASTAR Lasergene MegAlign 7.0 software. Taxonomic order adherence of 55 taxons identified in this study: (A) Lactobacillales; (B) Bacillales; (C) Fusobacteriales and Clostridiales; (D) Pseudomonadales; (E) Actinomycetales. Study isolates are shown in bold. Species in regular type were selected as (published) type strains of the different taxa. We used Escherichia coli K-12 rrnA [NCBI GenBank accession no. EG30084] and Mycobacterium tuberculosis H37Rv rrs [NC_000962] as outgroups.