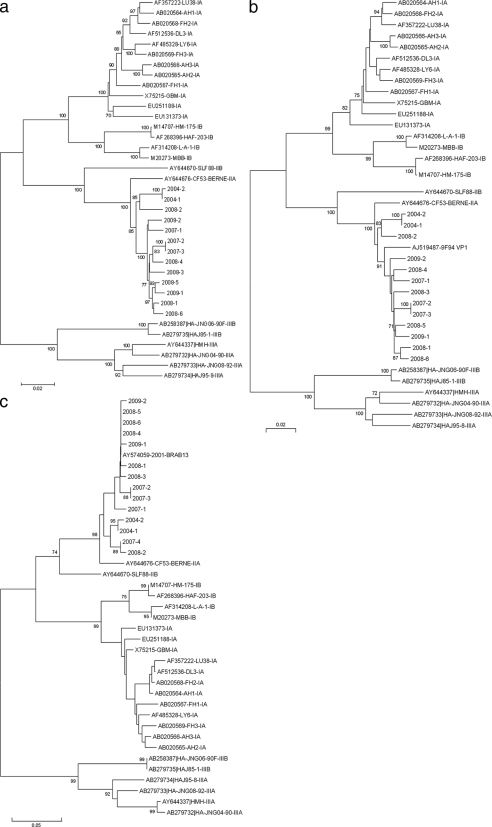
FIG. 3.
Phylogenetic analysis of clinical and reference isolates. Reference sequences are labeled with their GenBank accession numbers, strain codes, and defined subgenotypes according to the most recent classification of HAV isolates. Clinical isolates are labeled with their year of isolation and a number referring to the chronological order of isolation. Trees were constructed from a Kimura two-parameter genetic-distance matrix using the neighbor-joining method. (a) P1 sequences (2,295 nucleotides); (b) VP1 sequences (900 nucleotides); (c) VP1/2A sequences (486 nucleotides). Bootstrap values, obtained after 1,000 replications of bootstrap sampling, are given at the branches. Bars at the bottom of each phylogenetic tree denote distance.