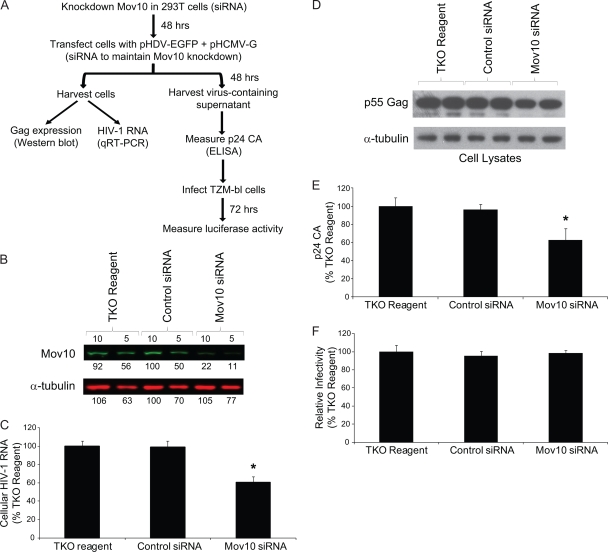
FIG. 4.
Effect of Mov10 knockdown on HIV-1 RNA and Gag expression, virus production, and virus infectivity. (A) Outline of experimental design. 293T cells were transfected in the absence of any siRNA (TKO reagent only), with a control siRNA, and with a Mov10 siRNA; 48 h later, pHDV-EGFP (1.0 μg) and pHCMV-G (0.2 μg) DNA and siRNAs were cotransfected into the cells in order to produce virus and maintain Mov10 knockdown. Total RNA was extracted from one fraction of the cells 48 h after transfection, and HIV-1 RNA (U5ψ target sequence) was quantified by real-time RT-PCR. Cell proteins from another fraction were analyzed by Western blotting using an anti-p24 CA antibody. The culture supernatants containing 5 ng of p24 CA were used to infect TZM-bl indicator cells, and luciferase activity was determined. (B) Knockdown of endogenous Mov10. Efficiency of Mov10 knockdown was assessed by quantitative Western blot analysis using an anti-Mov10 antibody 48 h after siRNA transfection. The protein input was normalized using α-tubulin. Different amounts (in μg; numbers shown above lanes) were loaded to facilitate protein quantitation, and the integrated signal intensities of the protein bands are shown relative to the control siRNA sample (set to 100%; numbers shown below each lane). (C) The knockdown of Mov10 reduces the steady-state levels of cellular HIV-1 RNA. PBGD RNA was used to normalize for the RNA input. (D) Knockdown of Mov10 decreases the intracellular steady-state levels of HIV-1 Gag. Cell lysates were analyzed by Western blotting using an anti-p24 CA antibody. The protein input was normalized using α-tubulin. (E) Effect of Mov10 knockdown on virus production. The amounts of virion released from cells were quantified by determination of p24 CA using ELISA. (F) Knockdown of Mov10 does not alter virus infectivity. Culture supernatants containing 5 ng of p24 CA were used to infect TZM-bl cells, and luciferase activity was determined. The average results from panels C, E, and F are shown relative to the sample with TKO reagent only (set to 100%). Error bars represent standard errors from four independent experiments. Asterisks indicate statistical significance (t test; P of <0.05).