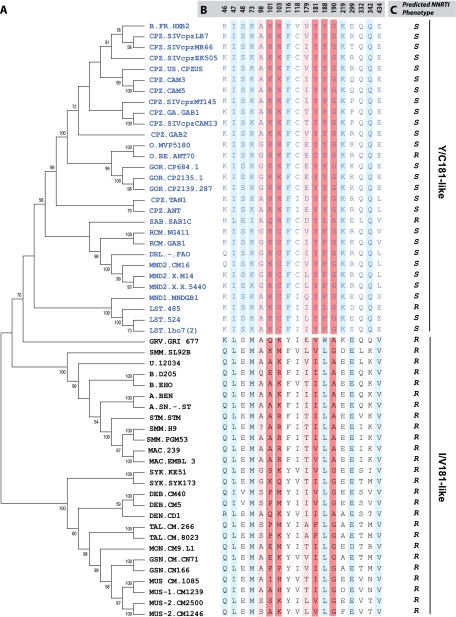
FIG. 1.
Phylogenetic analyses of primate lentivirus evolution in the reverse transcriptase gene. Phylogenetic analyses were performed with the RT of HIV and SIV strains (A). Sequences obtained from the HIV Los Alamos database were aligned to construct trees using neighbor-joining and Bayesian methods as described in Materials and Methods. The tree was bootstrapped with 1,025 repetitions. VESPA was used to identify conserved signature amino acid residues among SIV and HIV RT (B), leading to the identification of two distinct clusters, (Y/C181), which include all HIV-1 strains and a set of SIVs (SIVcpz, SIVmnd, SIVlst, SIVrcm), and (I/V181), which consists of HIV-2 and all monkey-derived SIVs. The numbers at the top of the alignment represent the RT amino acid positions. Amino acids known to confer high- and low-level resistance to NNRTIs are indicated in dark red and light red, respectively. Conserved positions linked to the Y/C181 or I/V181 lineages are indicated in sky blue. Linkage was performed using VESPA to identify conserved amino acid residues, i.e., those unique to the consensus sequence of each cluster. (C) Predicted NNRTI phenotypes are shown as R (resistant) or S (susceptible). Abbreviations for primates are as follows: CPZ, chimpanzees; HUM, humans; GOR, gorilla; RCM, red-capped mangabey; SAB, sabeus; DRL, drill; MND, mandrill; LST, l'Hoest monkey; GRV, grivet; SMM, sooty mangabey; STM, stump-tailed macaques; MAC, rhesus macaques; SYK, Sykes monkey; DEB, De Brazza monkey; TAL, talapoin; MON, mona monkey; GSN, greater spot-nosed monkey; MUS, mustached monkey.