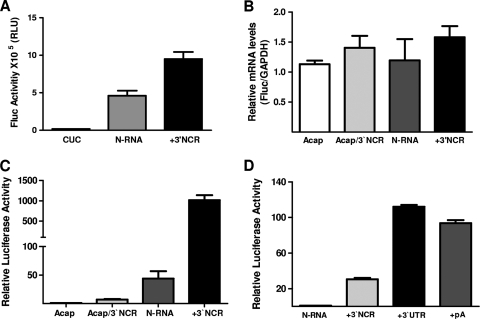
FIG. 2.
The 3′UTR of the ANDV cRNA synergistically stimulates cap-dependent translation initiation from virus-like mRNAs. (A) In vitro-transcribed N-RNA, CUC N-RNA (CUC), and N-RNA-3′NCR (+3′NCR) RNAs depicted in Fig. 1 were translated in RRL (50% [vol/vol]), and luciferase activity was measured as described in the text. FLuc activity is expressed as raw data. Values are the means ± standard deviations (SD) of results from three independent experiments, each conducted in triplicate. (B and C) In vitro-transcribed Acap-N-RNA (Acap), Acap-N-RNA-3′NCR (Acap/3′NCR), N-RNA, and N-RNA-3′NCR (+3′NCR) RNAs depicted in Fig. 1 were transfected into HeLa cells. (B) Total RNA was extracted from transfected cells and used in an RT-qPCR assay for FLuc and GAPDH as previously described (41). The relative mRNA content is expressed as FLuc/GAPDH. (C) Protein extracts from transfected HeLa cells were used to determine FLuc activity. FLuc activity for the Acap-N-RNA was arbitrarily set to 1 (±SD). Values are the means ± SD of results from three independent experiments, each conducted in triplicate. (D) The in vitro-transcribed N-RNA, N-RNA-3′NCR (+3′NCR), N-RNA-3′UTR (+3′UTR), and N-RNA-pA (+pA) RNAs depicted in Fig. 1 were transfected in HeLa cells together with mRNA in which the β-globin 5′UTR is followed by the Renilla luciferase (RLuc) reporter gene. Protein extract from transfected HeLa cells was used to determine FLuc and RLuc activities. FLuc values were normalized to the RLuc activities, and the relative luciferase activity (RLU) for the cap-N-RNA was arbitrarily set to 1 (±SD). Values are the means ± SD of results from three independent experiments, each conducted in triplicate.