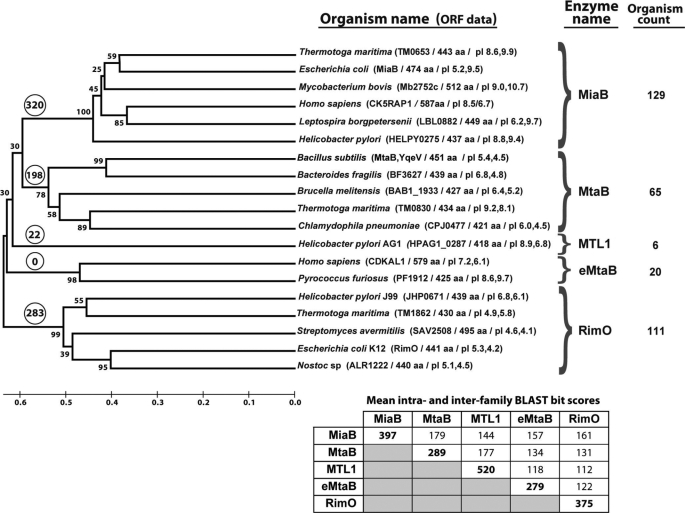
FIGURE 1.
Phylogenomic analysis of bacterial and human radical AdoMet methylthiotransferases. The cladogram shows the inferred evolutionary distances between representatives of all homologous protein families identified in a systematic search of the human genome and 474 fully sequenced bacterial genomes (see “Materials and Methods”). The MiaB and RimO families have been biochemically characterized in previous literature. This study described initial experimental characterization of representatives from the methylthiothreonylcarbamoyladenosine transferase B (MtaB) family and the eukaryotic methylthiothreonylcarbamoyladenosine transferase B (e-MtaB) family. The methYthiotransferase-like family 1 (MTL1), which is restricted to ϵ proteobacteria, has yet to be characterized experimentally. The e-MtaB family is found in archaebacteria but not eubacteria, although the other four families are found in eubacteria but not archaebacteria. The organism count indicates the number of unrelated bacterial genomes that encode a representative of each family (i.e. after correction for redundancy in genome organization). The numbers in circles at the root of each family indicate the number of times a member of one of the other families shown here is encoded simultaneously in the same genome (without redundancy correction). The numbers in a smaller font near the branch points indicate the percent of MEGA bootstrap replicates with the illustrated relative ordering of the successive branch points. Splits between the identified families confidently precede those within the families, with 100, 78, 98, and 99% confidence. Proper grouping of proteins in the MtaB family, which has the lowest 78% confidence for its first internal split, is supported by PSI-BLAST sequence profiling (data not shown) as well as the fact that proteins assigned to this family are never encoded in the same genome, even though 198 members of the other families are found encoded in the same genome as MtaB family members. The ORF data in parentheses includes the name of the protein, its amino acid sequence length, its overall pI, and the pI of its TRAM domain. The TRAM domains of bacterial MiaBs are generally basic, although those of bacterial RimOs are generally acidic. The TRAM domains of members of the other families show wider variations in pI values.