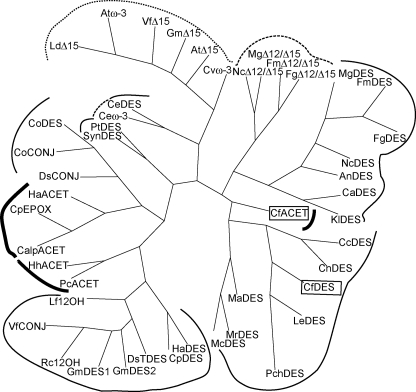
FIGURE 2.
Cladogram relating the chanterelle enzymes to representative FAD2, FAD3, and bifunctional desaturases. The graph shows the phylogenetic relationships between non-heme diiron enzymes diverged in chemo- and regioselectivity, assembled by a neighbor-joining algorithm from boot-strapped (n = 100) sequence alignments, plotted using DRAWGRAM (Phylip 3.65) (51). Families of desaturases and hydroxylases are bracketed by normal-weight lines, acetylenases by bold lines, enzymes with activity regioselective to the Δ15/ω-3 position by the dotted line, and enzymes with multiple locations of reactivity by the dashed lines. Chemoselectivity of the associated sequences are: DES, Δ12 desaturase; Δ12/Δ15, bifunctional Δ12/Δ15 desaturase; ACET, Δ12 acetylenase; CONJ, Δ12 conjugase; ω-3, ω-3 desaturase; ω-6, microsomal ω-6 desaturase; EPOX, 12 epoxygenase; 12-OH, 12-hydroxylase; TDES, trans- Δ12 desaturase; Δ15, Δ15 linoleoyl desaturase. The GenBankTM accession numbers for the displayed sequences are: AnDES, AAG36933; AtD15 (ER), P48623; Atw-3 chloroplast, P46310; CaDES, XP_722258; CalpACET, CAA76158; CcDES, AAU12575; CeDES, NP_502560; Cew-3, NP_001023560; CnDES, AAW42920; CoDES, AAG42259; CoCONJ, AAG42260; CpDES, CAA76157; CpEPOX, CAA76156; Cvw-3, BAB78717; DsCONJ, AAS72901; DsTDES, AAS72902; FgDES, EAA75859; FgD12/D15, ABB88516; FmDES, ABB88515; FmD12/D15, ABB88516; GmDES1, AAB00859; GmDES2, AAB00860; GmD15 (ER), P48625; HaACET, AAO38032; HaDES, AAL68982; HhACET, AAO38031; KlDES, XP_455402; LdD15, AAA86690; LeDES, BAD51484; Lf12OH, AAC32755; MaDES, BAA81754; McDES, BAB69056; MgDES, XP_365283; MgD12/D15, XP_362963; MrDES, AAM97924; NcDES, XP_959528; NcD12/D15, XP_329856; PcACET, AAB80697; PchDES, ACL13289; PtDES, AAO23564; Rc12OH, AAC49010; SynDES, NP_896789; VfCONJ, AAN87574; VfD15, AAC98967. An, Aspergillus nidulans; At, Arabidopsis thaliana; Ca, Candida albicans SC5314; Calp, C. alpina; Cc, Cryptococcus curvatus; Ce, Caenorhabditis elegans; Cn, Cryptococcus neoformans var. neoformans JEC21; Co, Calendula officinalis; Cp, Crepis palaestina; Cv, Chlorella vulgaris; Ds, Dimorphotheca sinuata; Fg, Gibberella zeae PH-1; Fm, Fusarium moniliforme; Gm, Glycine max; Ha, Helianthus annuus; Hh, Hedera helix; Kl, Kluyveromyces lactis; Ld, Limnanthes douglasii; Le, Lentinula edodes; Lf, Lesquerella fendleri; Ma, Mortierella alpina; Mc, Mucor circinelloides; Mg, Magnaporthe grisea 70-15; Mr, Mucor rouxii; Nc, Neurospora crassa OR74A; Pc, Petroselinum crispum; Pch, Phanerochaete chrysosporium; Rhizopus oryzae Pt, Phaeodactylum tricornutum; Rc, Ricinus communis; RoDES; Syn, Synechococcus sp. WH 8102; Vf, Vernicia fordii.