Abstract
The packaging of newly replicated and repaired DNA into chromatin is crucial for the maintenance of genomic integrity. Acetylation of histone H3 core domain lysine 56 (H3K56ac) has been shown to play a crucial role in compaction of DNA into chromatin following replication and repair in Saccharomyces cerevisiae. However, the occurrence and function of such acetylation has not been reported in mammals. Here we show that H3K56 is acetylated and that this modification is regulated in a cell cycle-dependent manner in mammalian cells. We also demonstrate that the histone acetyltransferase p300 acetylates H3K56 in vitro and in vivo, whereas hSIRT2 and hSIRT3 deacetylate H3K56ac in vivo. Further we show that following DNA damage H3K56 acetylation levels increased, and acetylated H3K56, which is localized at the sites of DNA repair. It also colocalized with other proteins involved in DNA damage signaling pathways such as phospho-ATM, CHK2, and p53. Interestingly, analysis of occurrence of H3K56 acetylation using ChIP-on-chip revealed its genome-wide spread, affecting genes involved in several pathways that are implicated in tumorigenesis such as cell cycle, DNA damage response, DNA repair, and apoptosis.
Keywords: Cell Cycle, Chromatin Histone Modification, DNA Damage, Gene Regulation, Histone Acetylase, Histone Deacetylase, p300, Sirtuins, DNA Damage Response (DDR)
Introduction
Genomic integrity is maintained by a complex interplay of DNA replication, repair, and checkpoint signaling. The compaction of eukaryotic DNA into chromatin affects these processes, and their mode of action in context of chromatin is not well understood (1, 2). Post-translational modifications (PTMs)6 of histones including acetylation, phosphorylation, methylation, ubiquitination, and sumoylation, which occur on their unstructured tails as well as in the globular core domains, control many aspects of chromatin function (3). Gene regulation and protection against DNA damage are among the important functions of histone tail modifications (4, 5). Several core domain modifications have been reported; however, the functions of very few have been studied in detail (6). Chromatin modifications establish a global chromatin environment in addition to orchestrating DNA-based processes (5). These modifications are believed to alter chromatin structure by affecting the recruitment of nonhistone proteins and interactions between nucleosomes and by disrupting chromatin contacts and are therefore implicated in regulation of various biological processes (7–9). It has also been proposed that histone modifications create a “histone code” that is utilized by nonhistone proteins targeting these proteins to sites of modification (10).
Acetylation of lysines of histones is most recognized for its ability to regulate gene expression; however, it has also been linked to replication and DNA damage tolerance (11–13). Acetylation is a dynamic process enabling its regulation at the level of both acetylation and deacetylation (11). Histones can be acetylated on multiple lysine residues, and their steady-state equilibrium is brought about by the opposing catalytic activities of histone acetyltransferases and histone deacetylases (14, 15). Recently, several groups have reported histone H3 core domain lysine 56 as novel site for acetylation in budding yeast Saccharomyces cerevisiae (16–18). This modification has been also reported to occur in the fission yeast, Schizosaccharomyces pombe (19) and in Drosophila (18). The unique feature of lysine 56 is its location in the nucleosome. It is the last residue of the αN-helix, which precedes the histone fold domain of histone H3, and it has been shown that histone-DNA interaction at the entry and exit points in the nucleosome weakens by its acetylation (16). Although some studies have indicated that H3K56 acetylation is involved in transcription, several groups have shown that it is also involved in the response to DNA damage during replication and nucleosome reassembly following repair and replication of DNA (15, 18, 20). In this study we monitored the presence of H3K56ac in mammalian cells and studied the enzymatic machinery involved in its regulation. We report that in mammals the p300 histone acetyltransferase acetylates H3K56, whereas hSIRT2 and hSIRT3 deacetylate H3K56ac. This acetylation is tightly regulated during cell cycle and peaks during the S phase of the cell cycle. We show that in response to DNA damage, acetylation of lysine 56 is up-regulated. Following DNA damage, the acetylated H3K56 that was diffused all over the nucleus relocalized to discrete nuclear foci that colocalize with double-strand break (DSB) markers γ-H2AX, phospho-ATM (pATM), and CHK2, which are localized at the site of damage repair. We also show that H3K56ac occurs in a genome-wide manner affecting multiple genes involved in cell cycle, DNA damage response, DNA repair, and apoptosis, among other pathways involved in tumorigenesis.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Treatment with DNA Damaging Agents
Cells were grown in DMEM (HEK 293T, HaCaT, and NIH 3T3), minimum Eagle's medium (HeLa), and RPMI 1640 medium (Jurkat and mouse thymocytes), supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. For transient transfections, the cells were seeded 24 h prior to transfection at 2 × 106 cells, and Lipofectamine 2000 (Invitrogen) was used for transfections according to the manufacturer's instructions. The cells were treated with methyl methane sulfonate (MMS, 0.02%) for 2 h, hydroxyurea (10 μm) for 24 h, camptothecin (2 μm) for 2 h, and γ irradiation (5 Gy) in a γ irradiation chamber (137Cs source).
Histone Preparation and Immunoblot Analysis
Histones were extracted and purified as described by Shechter et al. (21). Variable amounts of histone preparations (for different immunoblots) were resolved on 15% SDS-polyacrylamide gel and transferred to PVDF membrane, which was then probed with anti-pan H3 (Abcam; catalog no. ab1791), anti-H3K56ac (Upstate; catalog no. 07-677), anti-H3S10p (Upstate; catalog no. 05-806), anti-H3K9ac (Upstate; catalog no. 06-942), anti-pan H3ac (Upstate; catalog no. 06-599), and anti-γ-H2AX (Upstate; catalog no. 05-636) antibodies. The signal was detected by using VisualizerTM Western blot detection kit (Upstate; catalog no. 64-202).
Cell Synchronization
HeLa cells were synchronized at the G1/S phase boundary by double thymidine block. The cells were plated in minimum Eagle's medium at 30% confluency. At 60% confluency, 4 mm thymidine was added, and the cells were grown for 18 h. After 18 h, thymidine was washed off with sterile PBS, and minimum Eagle's medium supplemented with 10% fetal bovine serum was added. The cells were grown for 10–12 h before the addition of 4 mm thymidine again. After 18 h of incubation, thymidine was washed off to allow cells to progress into the cell cycle, and cells were harvested every 2 h (up to 12 h). The DNA content was analyzed by flow cytometry, and the level of H3K56ac was analyzed by Western blot using the above mentioned antibodies.
Cell Cycle Analysis
Cell cycle analysis was carried out by DNA content analysis using flow cytometry. The cells were fixed using ice-cold 70% ethanol, incubated for 2 h at −20 °C, and stained for 45 min with Guava cell cycle reagent containing propidium iodide. Cell cycle analysis was carried out in a Guava flow cytometer (Guava Easy Cyte; 0110-3660). The data were analyzed using Guava cell cycle cytosoft software (Guava Technologies). The level of H3K56 acetylation on cell synchronization and progression through the cell cycle was analyzed by immunoblot using the above mentioned anti-H3K56ac, anti-H3 antibodies, and anti-β-actin antibodies (Sigma).
In Vitro and in Vivo Acetylation
Histones extracted from HEK 293T cells were used for in vitro acetylation by p300. Briefly, histones were incubated with recombinant p300 (Upstate; catalog no. 14-418) using acetyl CoA as donor of acetyl group as per the procedure provided by the manufacturer. The reaction mixtures with and without p300 were resolved by SDS-polyacrylamide gel electrophoresis, and acetylated H3K56 was detected by immunoblot analysis. For in vivo acetylation assay p300 was transiently overexpressed by transfection of an expression construct (kind gift of Dr. A. Marcello). For knockdown of p300, HEK 293T cells were transfected with small interfering p300 duplex oligonucleotides (Santa Cruz) using siIMPORTER transfection reagent (Upstate; catalog no. 64-101) as per the manufacturer's instructions. The level of p300 expression was ascertained by immunoblotting with anti-p300 (Upstate; catalog no. 05-257).
Overexpression of Sirtuins and in Vivo Deacetylation
For in vivo deacetylation assay, human sirtuins hSIRT1-hSIRT7 were overexpressed in HEK 293T cells by transfection of different overexpression constructs (kind gifts of Dr. I. Horikawa) using Lipofectamine 2000 reagent (Invitrogen). Empty vector pCDNA 3.1 was used as a control. For siRNA-mediated knockdown of hSIRT2 and hSIRT3, HEK 293T cells were transfected with sihSIRT2 and sihSIRT3 (Qiagen) using transfection reagent supplied as per the manufacturer's instructions. The expression level of each sirtuin was monitored by immunoblot analysis using specific antibodies. The in vivo H3K56ac levels were monitored by immunoblot analysis.
Coimmunoprecipitation and Immunofluorescence Microscopy
Coimmunoprecipitation and immunofluorescence was carried out as described previously by Scully et al. (22). HeLa cells were grown on coverslips to 70% confluency and treated with 0.02% MMS for 2 h to induce DNA damage followed by three PBS washes. The cells were fixed in chilled methanol for 10 min, washed with PBS thrice, and blocked for 30 min in blocking solution (3% BSA in PBS). Next the cells were incubated with the following primary antibodies, diluted 1:100 in blocking solution for 1 h at room temperature as described in the figure legends: anti-H3K56ac (Upstate), anti-γ H2AX (Upstate), anti-CHK2 (A-11, SC17747; Santa Cruz Biotechnology, mouse monoclonal), anti-ATM kinase-S1981-P (Chemicon), and anti-p53 (Upstate). After two PBS washes, the coverslips were incubated with Alexa Fluor 488-conjugated donkey anti-rabbit and Alexa Fluor 647-conjugated goat anti-mouse secondary antibodies (Molecular Probes, Eugene, OR) for 1 h and washed with PBS thrice. The cells were stained with DAPI for 5 min and washed twice with PBS. The coverslips were mounted on slides using mounting medium (Sigma). Fluorescence images were captured using a confocal laser scanning microscope (Leica), and the data were analyzed using LCS software (Leica).
Immunoprecipitation (IP) and Determination of DNA Size in Input for IP
Coimmunoprecipitation was carried out as described previously by Scully et al. (22) with some modifications. 293T cells treated or untreated with MMS were pelleted down by centrifugation at 2000 rpm for 5 min after three PBS washes and trypsinization. Harvested cells were disrupted by four passages through a 25-gauge needle in 1 ml of 20 mm Tris/HCl, pH7.2, 1.5 mm KCl, 2.5 mm MgCl2, 0.05% Nonidet P-40, and protease inhibitor complex. Cell suspensions were centrifuged at 3000 × g for 5 min at 4 °C. After removal of supernatant, the nuclear pellets were resuspended in 0.5 ml of 50 mm Tris/HCl, pH 7.8, 150 mm KCl, 5 mm MgCl2, 250 mm sucrose, and protease inhibitor complex followed by five strokes of sonication at an output setting of 20 for 4 s. The nuclear extracts were then placed on top of a 0.5-ml cushion containing 0.88 m sucrose prepared in the same buffer as above for resuspending the nuclei followed by centrifugation at 13,000 × g for 15 min. This supernatant was used for immunoprecipitation using anti-γ-H2AX antibody. For DNA size determination, the supernatant was then treated with proteinase K (100 μg/ml) for 2 h at 50 °C and RNase for 30 min followed by phenol chloroform extraction. The DNA in the upper layer of phenol chloroform extraction was ethanol precipitated at −80 °C and recovered by centrifugation at 13,000 rpm for 20 min. The pellet was washed, air-dried, and dissolved in 50 μl of TE buffer; and DNA was then resolved by running on a 1% agarose gel.
Serum Starvation of Cells
The cells were serum-starved for G1 arrest essentially as described by Shin et al. (23). Briefly, HaCaT cells were grown to 70% confluency in DMEM supplemented with 10% fetal bovine serum (Invitrogen). The cells were then washed with plain DMEM and allowed to grow in serum-free medium for another 48 h before harvesting them for cDNA preparation and chromatin immunoprecipitation.
ChIP-on-chip and ChIP
A total of 1 × 108 Jurkat cells were treated with 10 mm sodium butyrate for 24 h and used for chromatin immunoprecipitation. ChIP was performed essentially as described by Kumar et al. (24). Briefly, formaldehyde cross-linked cells were sonicated to obtain fragments of genomic DNA ranging between 200 and 1000 bp. Sonicated chromatin was immunoprecipitated using anti-H3K56ac. Acetylated H3K56 associated chromatin was deproteinized and labeled using a chromatin labeling kit as per the manufacturer's instructions (Agilent Technologies). Input chromatin and H3K56ac ChIP sample were labeled with Cy3 and Cy5 dyes, respectively. These samples were hybridized to 2× 244,000 human tiling array (Agilent Technologies). The labeling, hybridization, scanning, and initial data analysis was carried out at Genotypic Technology (Bangalore, India). The data were analyzed using GeneSpring GX10 software (Agilent Technologies).
RESULTS
H3 Lysine 56 Is Acetylated and Up-regulated during S Phase of the Cell Cycle in Mammals
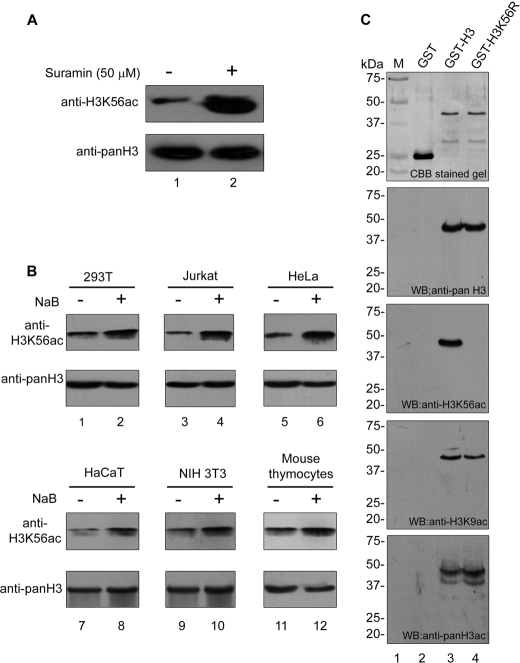
The histone H3 lysine 56 has been reported to be acetylated in yeast and Drosophila; however, it was not detected in mammals (18). A mass spectrometry study has reported very low levels of this modification in mammals (25). We hypothesized that the H3K56 may be acetylated transiently and therefore difficult to detect in mammals. Thus, it may be possible to detect this modification by incorporating two biochemical steps in the protocol: first enriching histones and second enriching the acetylation level of histones in general by treating cells with a histone deacetylase (HDAC) inhibitor. Because the class III NAD-dependent HDAC Hst3 and Hst4 were reported to deacetylate H3K56ac in yeast (19, 26), to test our hypothesis, we treated human cells with an inhibitor of NAD-dependent HDAC (class III), suramin, to shift the equilibrium toward acetylation. Immunoblot of histones from untreated and HDAC inhibitor-treated cells probed with antibodies specific to H3K56ac showed that indeed histone H3 is acetylated at lysine 56 in human cells, and this acetylation increases upon class III HDAC inhibition (Fig. 1A, top panel). Treatment of established human (HEK 293T, Jurkat, HeLa, and HaCaT) and mouse cell lines (NIH 3T3) as well as mouse thymocytes, with class I and class II HDAC inhibitor, sodium butyrate also resulted in increased H3K56 acetylation (Fig. 1B). These results show that H3K56 acetylation occurs in transformed cells (HEK 293T, lanes 1 and 2; Jurkat, lanes 3 and 4; HeLa, lanes 5 and 6), in normal cells (HaCaT, lanes 7 and 8; NIH 3T3, lanes 9 and 10), and in mouse thymocytes (Fig. 1B, lanes 11 and 12).
FIGURE 1.
Histone H3 is acetylated at lysine 56 in mammals. A, H3K56ac levels in HEK 293T cells in the absence (−) and presence (+) of NAD-dependent HDAC inhibitor suramin. HEK 293T cells were treated with suramin (50 μm, lane 2) for 24 h, and histones were enriched by acid extraction of nuclear proteins as described under “Experimental Procedures.” Cells without any treatment (lane 1) were taken as controls. Eight μg of acid extracted proteins were immunoblotted using anti-H3K56ac antibodies (upper panel). Total H3 detected by anti-pan H3 antibody was used as a loading control (lower panel). B, H3K56 acetylation increases in the presence of sodium butyrate. H3K56 acetylation levels in HEK 293T (lanes 1 and 2), Jurkat (lanes 3 and 4), HeLa (lanes 5 and 6), HaCaT (lanes 7 and 8), NIH 3T3 (lanes 9 and 10), and mouse thymocytes (lanes 11 and 12) in the absence (lanes 1, 3, 5, 7, 9, and 11) and presence (lanes 2, 4, 6, 8, 10, and 12) of 10 mm sodium butyrate (NaB) were monitored by immunoblotting. The cells were treated with 10 mm sodium butyrate for 24 h, and the histones were enriched by acid extraction of nuclear proteins as described under “Experimental Procedures.” 0.5 μg of acid extracted proteins were resolved by SDS-PAGE and immunoblotted using anti-H3K56ac (upper panels). Immunoblot using anti-pan H3 served as a loading control; however, to avoid signal saturation only 0.1 μg of acid extracted protein was used (lower panels). C, the specificity of anti-H3K56 ac antibody was determined by in vitro acetylation and immunoblotting. 250 ng each of GST (lane 2), GST-wtH3 (lane 3), and GST-mutH3K56R (lane 4) were resolved on a 10% SDS-PAGE gel (Fig. 1A; top panel). In vitro acetylation was performed with p300 as described under “Experimental Procedures,” and 50 ng of each sample was resolved on a 10% SDS-polyacrylamide gel. Immunoblotting was performed with anti-pan H3 (second panel), anti-H3K56ac (third panel), anti-H3K9ac (fourth panel), and anti-pan H3ac (fifth panel) antibodies. Molecular weight standards (lane 1) and the corresponding masses (in kDa) are indicated on the left for each panel. WB, Western blot.
Although the specificity of the anti-H3K56ac antibody used in this study has earlier been tested and reported by the authors using fission yeast H3K56R mutants (19), to further prove the specificity of this antibody, we have performed immunoblot analysis using wild type and mutant human H3 proteins. First, we cloned full-length human histone H3 in GST tag vector pC6-2 (27) to generate GST-H3 and also generated mutant H3 where the lysine residue at position 56 was replaced by an arginine residue (GST-H3K56R). Both the recombinant proteins were expressed in Escherichia coli and purified. To check the integrity of the recombinant proteins, 250 ng of each of GST (used as control), GST-H3, and GST-H3K56R was resolved on a 10% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue (Fig. 1C, first panel). All three proteins were then acetylated in vitro using recombinant p300 in the presence of acetyl CoA. Next 50 ng of each protein was resolved on a 10% SDS-polyacrylamide gel and immunoblotted with anti-pan H3 (Fig. 1C, second panel from top), anti-H3K56ac (Fig. 1C, third panel from top), anti-H3K9ac (Fig. 1C, fourth panel from top), and anti-pan H3ac (Fig. 1C, fifth panel from top). Immunoblot analysis using anti-pan H3 (Fig. 1C, second panel from top) revealed that equal amounts of wild type and mutant protein were used (compare lanes 3 and 4 in second panel). Immunoblot with anti-H3K56ac revealed that wild type GST-H3 was in vitro acetylated by p300 at lysine 56 (Fig. 1C, third panel from top, lane 3), whereas p300 failed to acetylate mutant GST-H3K56R at lysine 56 (Fig. 1C, third panel from top, lane 4). The immunoblots with anti-H3K9ac (Fig. 1C, fourth panel from top) and anti-pan H3ac (Fig. 1C, fifth panel from top) confirmed that the in vitro acetylation of GST-tagged proteins was successful. None of these antibodies could recognize the GST protein (Fig. 1C, all panels, lane 2).
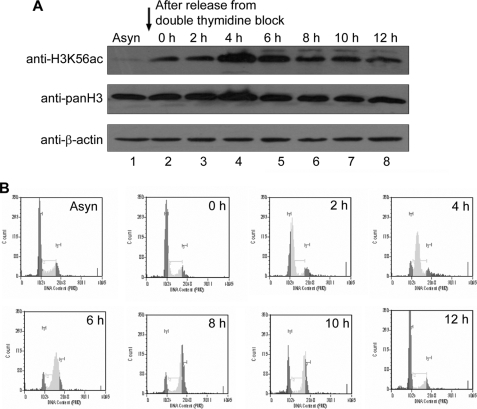
In yeast, H3K56ac occurs during the S phase and disappears during the G2/M phase during the normal cell cycle (16, 19, 26). We show that treatment of asynchronous HeLa cell population with sodium butyrate resulted in an increase in H3K56ac levels (Fig. 1B, top panel, lane 3). To determine whether the increase in acetylation on sodium butyrate treatment is due to an increase in S phase cells, flow cytometry analysis of sodium butyrate-treated HeLa cells was carried out. The flow cytometry analysis revealed that sodium butyrate treatment indeed resulted in an increase in S phase population (supplemental Fig. S1A). Therefore, it is possible that these acetylation levels vary during different phases of the cell cycle in mammals. To determine the H3K56 acetylation levels during different phases of the cell cycle, HeLa cells were synchronized at the G1/S boundary by double thymidine block, and then thymidine was washed out to enable cells to progress through the cell cycle. The cells were collected, before (Asyn) and after (0 h) thymidine treatment and subsequently every 2 h (up to 12 h) after release from arrest, and the level of H3K56 acetylation was monitored by immunoblot analysis as cells progressed through the cell cycle. Indeed histone H3K56 acetylation occurred in the synchronized cells, which are ready to enter the S phase (Fig. 2A). The acetylation peaked at 4 h (Fig. 2A, lane 4) when most of the cells are in the S phase, as can be inferred from the analysis of DNA content by flow cytometry (Fig. 2B and supplemental Fig. S1B). Therefore, these results suggest that H3K56 acetylation levels oscillate during the cell cycle and peak during the S phase, indicating a functional role for H3K56ac in S phase progression.
FIGURE 2.
H3K56 acetylation levels oscillate during cell cycle. A, HeLa cells were synchronized by double thymidine block and released to progress through the cell cycle as described under “Experimental Procedures.” The cells were collected before (Asyn) and after thymidine treatment (0 h) and every 2 h after release (up to 12 h); cell lysates were prepared; and H3K56 acetylation was analyzed by immunoblot. Immunoblotting with anti-pan H3 and anti-β-actin was used as a loading control. B, cell cycle progression was analyzed by monitoring DNA content by flow cytometry as described under “Experimental Procedures.” The dark gray, very light gray, and light gray peaks correspond to cells in the G1, S, and G2/M phases of the cell cycle, respectively.
Lysine 56 of Histone H3 Is Acetylated by p300 Acetyltransferase and Deacetylated by hSIRT2 and hSIRT3 Histone Deacetylases in Vivo
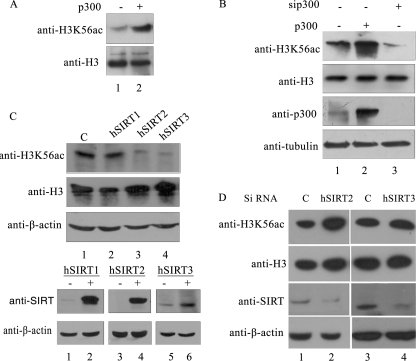
The H3K56 acetyltransferases and deacetylases are not known in mammals. The histone acetyltransferase Rtt109, which acetylates H3K56 in yeast (28, 29), is fungus-specific and does not share obvious homology with any protein in multicellular eukaryotes. However, the crystal structure of Rtt109 showed that there are structural similarities between Rtt109 and p300 in the histone acetyltransferase domain resulting from similar structural folds despite divergence in the amino acid sequence (30). This study indicated that fungal Rtt109 histone acetyltransferase could be a structural homolog of p300/CBP of higher organism. Therefore, to investigate whether p300 could potentially acetylate the histone H3 lysine 56, we performed in vitro acetylation by incubating acid extracted core histones from HEK 293T cells with recombinant p300 in the presence of acetyl CoA. The acetylation of H3K56 was promoted by p300 as evident from the increased signal for H3K56ac upon immunoblot analysis with anti-H3K56ac (Fig. 3A, lane 2). We next tested whether p300 acetylated H3K56 in vivo. Toward this, we overexpressed p300 and separately silenced it by RNA interference in HEK 293T cells. Overexpression of p300 resulted in increased acetylation of H3K56 (Fig. 3B, top panel, lane 2), whereas knockdown of p300 resulted in reduced acetylation of H3K56 (lane 3), confirming the direct role of p300 in acetylation of H3K56.
FIGURE 3.
Enzymes involved in the acetylation and deacetylation of H3K56 in human system. A, p300-mediated in vitro acetylation of histones extracted from HEK 293T cells. In vitro acetylation reaction was performed in the presence (lane 1) or absence (lane 2) of recombinant p300 as described under “Experimental Procedures.” The upper panel depicts immunoblot analysis using anti-H3K56ac antibodies, whereas the level of total H3 is shown in the lower panel. B, p300 acetylates H3K56 in vivo. p300 was overexpressed (lane 2) and silenced (lane 3) separately in HEK 293T cells as described under “Experimental Procedures.” The histones were isolated and immunoblotted for monitoring H3K56ac and H3 levels (upper two panels). Expression of p300 and tubulin was monitored by immunoblot analysis of the same samples. C and D, hSIRT2 and hSIRT3 deacetylate H3K56 in vivo. Sirtuins involved in the deacetylation of H3K56ac were tested by monitoring the reduction in the level of H3K56 acetylation upon overexpression of hSIRT1, hSIRT2, and hSIRT3 (C) and siRNA-mediated knockdown of hSIRT2 and hSIRT3 (D) by transient transfection in HEK 293T cells as described under “Experimental Procedures.” Immunoblotting anti-H3 and anti-β-actin was used as a loading control. Expression of the three sirtuins was confirmed using immunoblotting of lysates from control (−) and transfected (+) cells using anti-SIRT antibody (upper panel). Immunoblot with anti-β-actin (lower panel) was used as a loading control.
The H3K56 deacetylases Hst3 and Hst4 of budding yeast as well as Hst4 of S. pombe belong to the sirtuin family of NAD-dependent deacetylase, which has seven members in humans (19, 26). However, from the sequence homology of these seven sirtuins with that of yeast sirtuins, it is not clear which one is the functional homolog of these deacetylases. Therefore, to test whether any of the seven sirtuins is involved in deacetylation of H3K56, we overexpressed all of the seven human sirtuins in HEK 293T cells and checked how it effected acetylation of H3K56. Overexpression of only hSIRT2 and hSIRT3 resulted in a decrease in H3K56 acetylation (supplemental Fig. S1C and Fig. 3C, lanes 3 and 4). There was no decrease in the H3K56ac level on overexpression of other sirtuins, clearly indicating that hSIRT2 and hSIRT3 are responsible for deacetylating H3K56 in vivo in mammalian cells. Knockdown of hSIRT2 and hSIRT3 by siRNA resulted in increased acetylation of H3K56 (Fig. 3D), further confirming their role in deacetylation of H3K56.
Histone H3 Lysine 56 Is Hyperacetylated in Response to DNA Damage
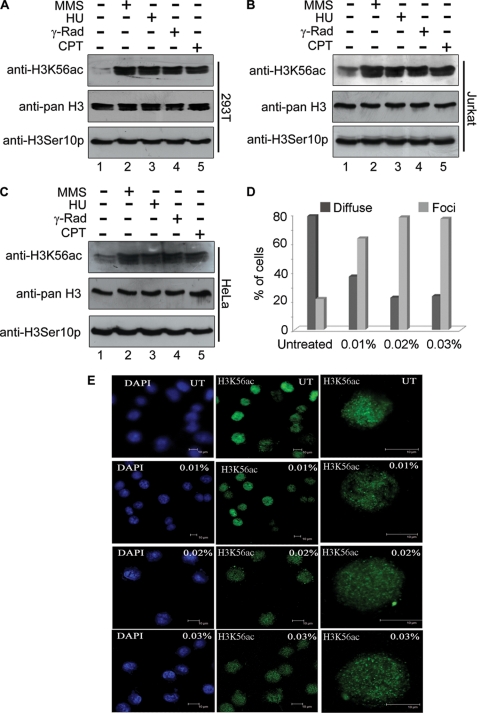
Although the studies in yeast have provided valuable clues to the function of histone H3 lysine 56 acetylation, in mammals its function has not been explored. Given the potential role of this acetylation in DNA damage response, we examined whether its levels are altered on DNA damage. To determine the levels of H3K56 acetylation in response to DNA damage, HEK 293T, Jurkat, and HeLa cells were treated with various DNA-damaging agents such as MMS, hydroxyurea, γ-radiation and camptothecin. Subsequently, histones were extracted and analyzed by immunoblot with anti-H3K56ac. The results of this experiment show that the levels of H3K56 acetylation increase when cells encounter DNA-damaging agents (Fig. 4, A–C, upper panels, lanes 2–5) in all three cell types. There was no change in the level of H3 serine 10 phosphorylation observed upon DNA damage (Fig. 4, A–C, lower panels). Collectively, these results suggest that the increased level of H3K56 acetylation in mammalian cells seems to be required for cellular response to DNA damage. This also indicates that the role of this acetylation may be functionally conserved in mammals.
FIGURE 4.
DNA damage-induced hyperacetylation and relocalization of histone H3 lysine 56 to subnuclear foci. A–C, H3K56ac levels increased in HEK 293T cells (A), Jurkat cells (B), and HeLa cells (C) on treatment with various DNA-damaging agents, viz. MMS (0.015%, lanes 2), hydroxyurea (10 μm, lanes 3), γ-irradiation (5 grays, lanes 4), and campetothecin (2 μm, lanes 5). (−) and (+) indicate the absence and presence, respectively, of the DNA-damaging agents. The cells were treated with various DNA-damaging agents as described under “Experimental Procedures,” and histones were then prepared by acid extraction. Immunoblot analysis was performed using anti-H3K56ac (top panel), anti-pan H3 (middle panel), and anti-H3S10p (bottom panel). D and E, DNA damage-dependent relocalization of H3K56 acetylation to discrete nuclear foci. HEK 293T cells were treated with different concentrations of MMS as indicated on the top right corner of each panel on the left (E). The cells were then immunostained as described under “Experimental Procedures.” The first two columns of panels in E show fields of multiple cells immunostained with anti-H3K56ac, and DNA was counterstained with DAPI. The last column of panels shows higher magnification images of single cells immunostained with anti-H3K56ac antibody. D, the percentages of cells showing diffused staining and discrete foci are shown; in all samples, 250 cells were counted and plotted in a bar graph.
Acetylated H3 Lysine 56 Forms Nuclear Foci in Response to DNA Damage
Cells activate checkpoint and DNA repair pathways in response to DNA damage. Proteins involved in DNA damage checkpoint, DNA repair, and chromatin remodeling form DNA damage-regulated aggregates that are cytologically detectable as subnuclear foci in the cells (7, 29, 32). To determine whether acetylation of H3K56 participates in DNA damage-signaling pathways, we examined localization of acetylated H3K56 upon MMS-induced DNA damage, by immunofluorescence analysis using anti-H3K56ac. Strikingly, acetylated H3K56 that was spread diffusely throughout the nuclei of untreated cells relocated to discrete subnuclear foci in response to MMS treatment (Fig. 4E). The number of foci also increased with increasing concentrations of MMS, suggesting that this was a direct response to DNA damage imparted by MMS. Quantification of the number of foci from 250 cells for each treatment revealed that the number of foci increased with increasing concentrations of MMS (Fig. 4D).
Acetylated H3K56 Colocalizes with γ-H2AX in Response to DNA Damage
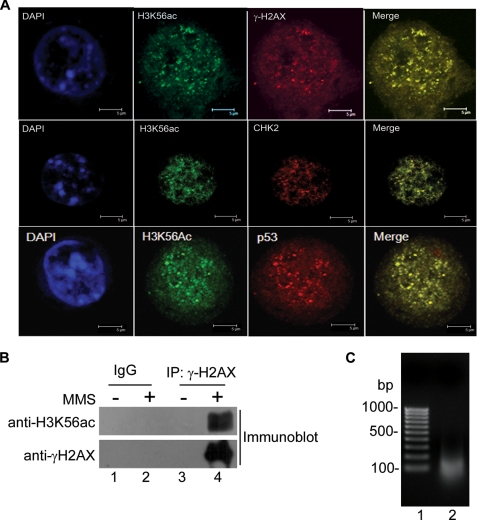
The foci formed by acetylated H3K56 in response to DNA damage (Fig. 4E) appeared very similar to those reported for phosphorylated H2AX (γ-H2AX) (31, 32). H2AX is one of the variants of histone H2A in mammalian cells that becomes rapidly phosphorylated when cells encounter DNA damage (29, 32). Phosphorylated H2AX appears at the site of DNA damage as discrete nuclear foci within a short interval after DNA damage (31). To examine whether the acetylated H3K56 is also present at the sites of DNA damage along with γ-H2AX, we checked by immunofluorescence analysis whether they colocalize. As shown in Fig. 5A (top row, lane 4), damage-induced acetylated H3K56 foci completely colocalized with γ-H2AX foci. Because upon DNA damage, H3K56 acetylation is also increased and colocalized with γ-H2AX, it is highly likely that acetylated H3K56 tend to become incorporated in the same nucleosome at the site of DNA damage. We therefore argued that these two could be then coimmunoprecipitated from cells that have been insulted with DNA-damaging agents. Indeed, antibody to γ-H2AX coimmunoprecipitated H3K56ac from extracts of MMS-treated HEK 293T cells (Fig. 5B, lane 4). We next wanted to determine whether both γ-H2AX and histone H3 acetylated at Lys56 exist within the same nucleosome. To investigate whether γ-H2AX and H3K56ac are in the same nucleosome, DNA was isolated from the input material used for coimmunoprecipitation as described under “Experimental Procedures,” and its size was determined by resolving it on a 1% agarose gel. As can be seen from Fig. 5C, the size of the majority of DNA in the input material is between 100 and 200 base pairs, indicating that it mostly contained mononucleosomes. Thus, immunoprecipitation analysis revealed that H3K56ac and γ-H2AX biochemically interact after MMS treatment, suggesting that both of these are likely to exist within the same nucleosome at the site of DNA damage. In addition, to test whether H3K56ac colocalizes with other proteins activated by DNA damage, we performed colocalization of H3K56ac with pATM, CHK2, and p53, which are well known markers of DSBs. The H3K56ac foci colocalized with pATM, Chk2, and p53 foci (Fig. 5A and supplemental Fig. S2B). Coimmunostaining of these DSB markers with H3K56ac in untreated HEK 293T cells is shown in supplemental Fig. S2. Only secondary antibody controls were also carried out to rule out the possibility of cross-reactivity and channel bleed through (supplemental Fig. S1D). Our data demonstrate that acetylated H3K56 colocalizes with several markers of DNA damage and interacts with γ-H2AX at the sites of DNA double-strand breaks upon MMS treatment.
FIGURE 5.
Localization of acetylated H3K56 at the sites of DNA repair. A, HEK 293T cells were treated with 0.02% MMS and coimmunostained with anti-H3K56ac antibody and anti-γ-H2AX antibody (top panels), anti-Chk2 antibody (middle panels), and anti-p53 antibody (bottom panels). The nuclei were stained with DAPI. B, coimmunoprecipitation of H3K56ac and γ-H2AX on DNA damage. HEK 293T cells were treated with 0.02% MMS, and the cell lysates were prepared as described under “Experimental Procedures.” Coimmunoprecipitation was carried out with anti-γ-H2AX antibody. The immunoprecipitated samples were run on 15% SDS-PAGE and immunoblotted using anti-H3K56ac antibodies and anti-γ-H2AX antibodies as indicated. C, size of DNA in the lysates used for IP. Agarose gel (1%) showing the size of DNA fragment in the input for IP (lane 2). DNA was isolated from lysates prepared for IP as described under “Experimental Procedures” and run on a 1% agarose gel.
Acetylation of H3K56 Occurs Genome-wide and Also Preferentially Marks the Genes Involved in DNA Damage Response Pathways
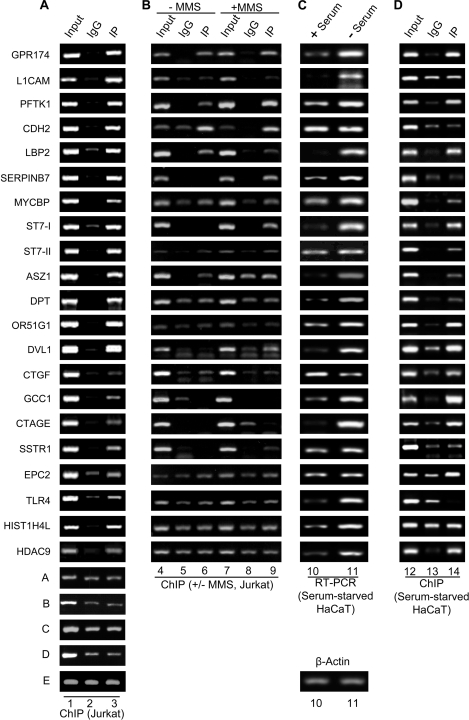
Immunofluorescence analysis suggested a genome-wide occurrence of H3K56ac in the human cells. To further ascertain whether this acetylation was restricted to certain regions of the genome or scattered throughout, we performed analysis of genome-wide occupancy of H3K56ac using ChIP-on-chip. Chromatin was immunoprecipitated from Jurkat cells after treating the cells with 10 mm sodium butyrate for 24 h, and ChIP was performed as described under “Experimental Procedures.” Input and immunoprecipitated chromatin was labeled with Cy3 and Cy5 dyes, respectively, and hybridized to 2 × 244,000 human tiling array, which represented most of the human genes and intergenic regions. We found that 12,927 probes (∼3%) showed enrichment of 2-fold or more for H3K56ac (Fig. 6). Because each gene was represented by multiple probes, these probes correspond to 4,504 genes that were enriched 2-fold or more in the H3K56ac immunoprecipitated chromatin from Jurkat cells. These probes mapped along the lengths of all chromosomes with a few dense clusters revealed by chromosome-wise position analysis of the enriched probes (Fig. 6). To confirm that the fold occupancy values obtained by ChIP-on-chip analysis were real representations of the in vivo occupancy, we performed validation experiments for the occupancy of H3K56ac on some of the key genes involved in diverse cellular pathways. For this, we designed primers corresponding to the genomic regions flanking the probes enriched in the ChIP-on-chip analysis and used them for PCR amplification of the DNA purified from chromatin immunoprecipitated with anti-H3K56ac after treatment of Jurkat cells with sodium butyrate for 24 h. The ChIP-PCRs revealed that occupancy of H3K56ac was enriched multiple-fold on many transcriptionally hyperactive genes, and those probes that were negative in the ChIP-on-chip analysis did not show any significant enrichment over IgG (Fig. 7A, compare lane 3 with lane 2). Quantitative ChIP-PCRs confirmed the enhanced occupancy of H3K56ac at these regions (supplemental Table S1). Interestingly, pathway analysis revealed that genes enriched in H3K56ac occupancy were involved in diverse cellular pathways (supplemental Table S2); among these were several genes important for cell cycle regulation (supplemental Fig. S4), DNA repair (supplemental Fig. S5–S7), and apoptosis (supplemental Fig. S8). To evaluate the effect of DNA damage (MMS treatment) on the occupancy of H3K56ac on multiple gene loci, we performed ChIP for a few representative genes using the chromatin immunoprecipitated from control Jurkat cells and Jurkat cell treated with 0.02% MMS for 2 h. It was revealed that most of these genes showed increased H3K56ac occupancy upon DNA damage induced by MMS treatment (Fig. 7B, compare lane 9 and lane 6), which is consistent with our Western blot data. Few genes such as MYCBP, DPT, ORI51G1, and HIST1H4L did not show any enrichment in treated cells over untreated cells, indicating the role of alternative mechanism(s) in the regulation of these genes. Further, to monitor the transcript levels of genes involved in DNA damage response and cell cycle, Jurkat cells were treated with increasing concentrations of MMS (0.005%, lane 2; 0.01%, lane 2; 0.02%, lane 3; and 0.04%, lane 4) for 2 h, and RT-PCR was performed for a few representative genes (supplemental Fig. S9). Transcription of some of these genes increased on MMS treatment, but a few did not show significant change. The increase in H3K56ac occupancy at the genes where it is enriched could be linked to replication or transcription. To clearly distinguish H3K56Ac linked to transcription from replication, HaCaT cells (normal human keratinocytes) were arrested in the G1 phase of cell cycle, and the transcriptional status of these genes was tested. HaCaT cells were serum-starved for 36 h to arrest them at G1 stage, and cDNA was prepared from control and serum-starved cells. Quantitative RT-PCRs were then performed for the corresponding genes. Interestingly, transcript levels of most of the genes increased upon serum starvation of the cells (Fig. 7C, lanes 10 and 11, and supplemental Table S1). The CT values for all the genes were normalized with that of β-actin (Fig. 7C, bottom panel), and the fold change was calculated for each gene. However, it was observed that some of the genes (e.g. CDH2, CTGF, SSTR1, and EPC2) showed similar or lesser transcriptional activity as compared with the nonstarved cells. To further strengthen the fact that H3K56ac is present on the genes involved in various cellular processes, we performed ChIP analysis of chromatin isolated from HaCaT cells, which were serum-starved and treated with 10 mm sodium butyrate for 24 h, using anti-H3K56ac. Quantitative ChIP-PCR analysis revealed that most of the genes showed significant enrichment in chromatin immunoprecipitated with anti-H3K56ac (Fig. 7D, lane 14, and supplemental Table S1) as compared with that with IgG (lane 13). Certain genes did not show a significant increase in immunoprecipitated chromatin, and future studies will be required for addressing this “aberrant” pattern of some of the genes. Taken together, these results provide further indication in support of our finding that H3K56 acetylation functions in the DNA damage response in human cells.
FIGURE 6.
Genome-wide distribution of H3K56 acetylation. Map depicting genome-wide occupancy of H3K56 acetylation. ChIP-on-chip was performed using anti-H3K56ac antibody as described under “Experimental Procedures,” and analysis of enriched genomic regions was carried out using Genespring GX10 software (Agilent Technologies). Each vertical line on the chromosomal map represents the location of an enriched probe. Certain regions on various chromosomes show clustering of H3K56ac occupancy.
FIGURE 7.
H3K56ac is enriched in upstream regulatory regions of multiple genes involved in the cellular pathways implicated in DNA damage response and tumorigenesis in Jurkat cells and G1 arrested HaCaT cells. A, ChIP analysis was performed in Jurkat cells treated with 10 mm sodium butyrate for 24 h using anti-H3K56ac antibody, as described under “Experimental Procedures.” The gel pictures in individual panels depict PCR-amplified products using primers specific for the indicated genes and the DNA eluted from chromatin, immunoprecipitated using anti-H3K56ac (lane 3) and control rabbit IgG (lane 2). Lane 1, input chromatin. The last five panels (rows A–E) depict the controls from genomic loci that show no specific enrichment in the immunoprecipitated chromatin over the control IgG. B, ChIP analysis was performed in control (− MMS, lanes 4–6) Jurkat cells and Jurkat cells treated with 0.02% MMS for 2 h (+ MMS, lanes 7–9) as described under “Experimental Procedures.” The gel pictures in individual panels depict the PCR-amplified products using primers specific for the indicated genes and the DNA eluted from chromatin, immunoprecipitated using anti-H3K56ac (lanes 6 and 9) and control rabbit IgG (lanes 5 and 8). Lanes 4 and 7 depict input chromatin. C, real time quantitative RT-PCR was performed for monitoring the expression levels of the corresponding genes from A and B using cDNA obtained from control (+ serum, lane 10) and G1-arrested, serum-starved (− Serum, lane 11) HaCaT cells. The CT values for various genes were normalized with that of β-actin (bottom panel). D, ChIP was performed using anti-H3K56ac antibody in serum-starved HaCaT cells treated with 10 mm sodium butyrate for 24 h. The gel pictures in the individual panels depict PCR-amplified products using primers specific for the indicated genes and the DNA eluted from chromatin, immunoprecipitated using anti-H3K56ac (lane 14) and control rabbit IgG (lane 13). Lane 12, input chromatin. Quantitative ChIP-PCRs were performed, and the fold enrichment over IgG was calculated as described under “Experimental Procedures.” The quantification of all of the above PCRs is tabulated in supplemental Table S2.
DISCUSSION
Our studies demonstrate that histone H3 lysine 56 is acetylated in mammalian cells and functions in the DNA damage response pathway. Several histone modifications have been identified so far that contribute to cellular response to DNA damage, including histone H3 lysine 56 (5). However, earlier studies were unable to detect H3K56 acetylation in mammalian cells (18). We show H3K56 acetylation for the first time along with three other groups in human cells (33–35). Our studies show that inhibition of both NAD-dependent HDAC as reported earlier in yeast (19, 26) and NAD-independent HDAC (class I and II) resulted in increased levels of H3K56 acetylation in mammals (Fig. 1, A and B). The increase in acetylation of histone H3K56 by inhibitors of class I and class II HDACs indicates that either this lysine could be targeted by these classes of HDACs, or it could also be due to differential regulation of expression of sirtuins by these classes of HDAC inhibitors (36). We have, for the first time, demonstrated conclusively, the specificity of anti-H3K56 antibody used in this and many previous studies, by performing various immunoblotting experiments with recombinant wild type GST-H3 and mutant GST-H3K56R proteins (Fig. 1C). We demonstrate that H3K56 acetylation is up-regulated during the S phase of the cell cycle in mammalian cells (Fig. 2A). Our data demonstrate that acetylated H3 lysine 56 has a role during the normal S phase of the cell cycle. This aspect of H3K56ac function will be investigated in detail in the future with respect to its role in DNA replication and interaction with replication proteins. In yeast, H3K56 acetylation is known to occur predominantly on newly synthesized histones that are assembled into chromatin after DNA replication (16) and are rapidly deacetylated after the S phase (19, 26, 37). This acetylation and deacetylation of H3K56 is regulated during the cell cycle and presumably by the DNA damage checkpoint. Nevertheless, the acetylation persists during DNA damage repair because the deacetylases Hst3 and Hst4 that belong to the sirtuin family are tightly regulated such that they are expressed after the S phase but repressed upon checkpoint activation (17, 26).
Furthermore, we report that the histone acetyltransferase p300 acetylates H3K56, and the human sirtuins hSIRT2 and hSIRT3 deacetylate it in vivo in human cells (Fig. 3). In yeast, Rtt109 acetylates H3K56, and Hst3 and Hst4 deacetylate it. Rtt109 needs either of the two histone chaperones, Asf1 or Vsp75, for enzymatic activity in vivo (38, 39). Recent studies have shown that acetylated H3K56 is incorporated into replicating DNA and enhances the binding affinity of CAF1 and Rtt106 for histone H3, which is essential for replication-coupled nucleosome assembly. Interestingly, the in vitro acetylation reaction (Fig. 3A) was performed in the absence of histone chaperones, suggesting that they are not obligatory for the acetylation of H3K56 in mammalian systems. However, the possibility of their requirement for damage-inducible modification and subsequent DNA repair cannot be ruled out and needs to be tested further.
Discovery of enzymatic machinery involved in the acetylation-deacetylation of H3K56 provides important clues toward elucidation of the functional significance of H3K56 acetylation in mammalian cells. It also remains to be established which of the seven human sirtuins (hSIRT1–7) is a functional homolog of Hst3 and Hst4 deacetylases of yeast. Our discovery of hSIRT2 and hSIRT3 as deacetylases for H3K56 (Fig. 3C) provides functional candidates for such analysis. It will be very interesting to delineate whether the functional homolog of yeast Hst4 in humans also exhibits similar functions in DNA damage response and, if so, to determine its contributions toward manifestation of diseases such as cancer. Moreover, hSIRT2 has been known as a cytoplasmic protein; later on it was also shown to play a role in mitosis (40). Vaquero et al. (41) have also shown it to be a histone H4 Lys16 deacetylase. Our report also provides further support for hSIRT2 having a nuclear function. This aspect of SIRT2 function requires further investigation. Similarly, the localization of hSIRT3 is controversial, with most studies indicating it to be a mitochondrial NAD-dependent deacetylase. However, recently it has also been shown to deacetylate histone H4 Lys16 (42). Our data provide further evidence for hSIRT3 having nuclear function as it deacetylates histone H3K56ac (Fig. 3C).
Our results also show the up-regulation of H3K56 acetylation on treatment of cells with genotoxic agents such as MMS, hydroxyurea, γ-irradiation, and camptothecin (Fig. 4A-C). When a cell encounters DNA damage, acetylated H3K56 that was diffused throughout the nucleus relocalizes to discrete nuclear foci (Fig. 4D and E). Furthermore, in the presence of DNA damage, H3K56ac colocalizes with DNA damage marker γ-H2AX at the sites of DNA repair (Fig. 5A), and both proteins interact biochemically, as revealed by coimmunoprecipitation (Fig. 5B). Thus, DNA damage triggers the recruitment of acetylated H3K56 into the discrete nuclear foci, presumably via its incorporation into the nucleosomes at the site of DNA damage. Our study shows that acetylation of H3K56 could be an integral part of the γ-H2AX/ATM/Chk2 pathway regulating downstream effectors like tumor suppressor p53, with which it also colocalizes (Fig. 5A). It has been shown in clinical specimens from patients with various stages of tumors that they have increased levels of marker proteins involved in DNA damage signaling pathways such as γ-H2AX, pATM, and CHK2 (43). We hypothesize that like γ-H2AX, acetylated H3K56 is a potential marker for tumorigenesis. It will be interesting to study the levels of H3K56ac during tumor initiation and progression using samples from patients with tumors at different stages of tumorigenesis. Indeed a very recent report has shown that acetylation of H3K56 is increased in multiple types of cancer, correlating it with increased levels of the histone chaperone ASF1A in these tumors (33).
Another important outcome of these studies is the identification of genome-wide targets of this histone modification. ChIP-on-chip studies indicated that although H3K56 acetylation is distributed on all chromosomes, it seems to occur preferentially at certain loci (Fig. 6). Moreover, the genes enriched in H3K56ac occupancy represented diverse cellular pathways such as cell cycle regulation, DNA repair, and apoptosis (Fig. 7, supplemental Tables S1 and S2 and supplemental Figs. S4–S8). In yeast, acetylation of H3K56 is known to preferentially occur at transcriptionally hyperactive genes such as histones (18). Chromatin immunoprecipitation assay revealed that H3K56ac levels increased at the above mentioned gene loci upon treatment of cells with MMS (Fig. 7 B). We have also observed enrichment of H3K56ac at histones genes in addition to multiple other genes from the above mentioned pathways, indicating that a similar mechanism may act at the level of most genes that contain acetylated H3K56. It was also observed that these genes were actively transcribed and showed H3K56ac enrichment when normal diploid human cells (HaCaT) were serum-starved to arrest them in the G1 stage of the cell cycle (Fig. 7C and D). This is a crucial observation pointing toward the fact that acetylation at H3K56 is an important modification that is enriched in the G1 stage and that is required for proper progression of the cell cycle. However, further investigation is required to address the molecular mechanism of how H3K56 acetylation contributes to increased transcription of genes.
The significance of covalent modification of histones in damage response, DSB repair, and checkpoint signaling has become a subject of intense study recently (7, 8). A major challenge in this area, at present, is to uncover the molecular determinants of the recruitment of specific components of the remodeling machinery at the DNA lesions and to define how histone-modifying enzymes interact with core components of the DNA damage surveillance and repair machinery. These studies are also important for elucidation of the molecular mechanisms of how chromatin modifications influence specific biochemical steps in the DNA damage response pathway. Our study reports the occurrence of acetylation of H3 core domain lysine 56 and its function in the DNA damage response pathway in human cells for the first time, and it could have very important implications in cancer initiation and progression.
In mammals, phosphorylation of H2AX (γ-H2AX) has been identified as a PTM associated with DNA DSBs. According to histone code hypothesis (10), there may be additional PTMs at DSBs that could synergize with γ-H2AX, and later on it was shown that H2B serine 14 phosphorylation foci are dependent on γ-H2AX at the DSB to facilitate damage repair (7, 44). It remains to be investigated whether any other PTM is also involved in this process. Furthermore, it will be interesting to study the role of H3K56 acetylation in mammals to determine whether it is dependent on the phosphorylation of H2AX. Unique combinations of PTMs result in subtle changes in the packaging theme that provide “instructions” as to how the DNA template is to be read when required, often leading to complex modification patterns that correlate with various states of gene expression or other DNA-templated processes. However, the mechanisms by which these multiple chemical marks might be translated into meaningful biological responses are not well understood. Because of the identification of the enzymatic machinery involved in this unique, reversible modification and demonstration of its genome-wide occurrence, this study ultimately holds promise for predictive, better diagnostics and therapeutic measures for treating disorders, such as cancer, associated with genomic instability.
Supplementary Material
Acknowledgments
We thank Dr. A. Marcello for p300 expression construct and Dr. I. Horikawa for sirtuin expression constructs. We also thank C. S. Murthy and M. Nalini of University of Hyderabad for helping with the confocal microscopy.
This work was supported by a grant from the Institute of Life Sciences (to D. H.) and grants from the Department of Biotechnology of the Government of India and the Wellcome Trust (to S. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S9.
- PTM
- post-translational modification
- DSB
- double-strand break
- MMS
- methyl methane sulfonate
- IP
- immunoprecipitation
- HDAC
- histone deacetylase.
REFERENCES
- 1.Ehrenhofer-Murray A. E. (2004) Eur. J. Biochem. 271, 2335–2349 [DOI] [PubMed] [Google Scholar]
- 2.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 3.Peterson C. L., Laniel M. A. (2004). Curr. Biol. 14, R546–R551 [DOI] [PubMed] [Google Scholar]
- 4.Bird A. W., Yu D. Y., Pray-Grant M. G., Qiu Q., Harmon K. E., Megee P. C., Grant P. A., Smith M. M., Christman M. F. (2002) Nature 419, 411–415 [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove M. S., Boeke J. D., Wolberger C. (2004) Nat. Struct. Mol. Biol. 11, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 7.Downs J. A., Nussenzweig M. C., Nussenzweig A. (2007) Nature 447, 951–958 [DOI] [PubMed] [Google Scholar]
- 8.Groth A., Rocha W., Verreault A., Almouzni G. (2007) Cell 128, 721–733 [DOI] [PubMed] [Google Scholar]
- 9.Suganuma T., Workman J. L. (2008) Cell 135, 604–607 [DOI] [PubMed] [Google Scholar]
- 10.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 11.Kurdistani S. K., Grunstein M. (2003) Nat. Rev. Mol. Cell Biol. 4, 276–284 [DOI] [PubMed] [Google Scholar]
- 12.Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., Yates J. R., 3rd, Abmayr S. M., Washburn M. P., Workman J. L. (2004) Science 306, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 13.van Attikum H., Gasser S. M. (2005) Nat. Rev. Mol. Cell Biol. 6, 757–765 [DOI] [PubMed] [Google Scholar]
- 14.Lee K. K., Workman J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 15.Miller K. M., Maas N. L., Toczyski D. P. (2006) Cell Cycle 5, 2561–2565 [DOI] [PubMed] [Google Scholar]
- 16.Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 17.Ozdemir A., Spicuglia S., Lasonder E., Vermeulen M., Campsteijn C., Stunnenberg H. G., Logie C. (2005) J. Biol. Chem. 280, 25949–25952 [DOI] [PubMed] [Google Scholar]
- 18.Xu F., Zhang K., Grunstein M. (2005) Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 19.Haldar D., Kamakaka R. T. (2008) Eukaryot. Cell 7, 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rufiange A., Jacques P. E., Bhat W., Robert F., Nourani A. (2007). Mol. Cell 27, 393–405 [DOI] [PubMed] [Google Scholar]
- 21.Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 22.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M. (1997) Cell 88, 265–275 [DOI] [PubMed] [Google Scholar]
- 23.Shin J. S., Hong S. W., Lee S. L., Kim T. H., Park I. C., An S. K., Lee W. K., Lim J. S., Kim K. I., Yang Y., Lee S. S., Jin D. H., Lee M. S. (2008) Int. J. Oncol. 32, 435–439 [PubMed] [Google Scholar]
- 24.Kumar P. P., Purbey P. K., Ravi D. S., Mitra D., Galande S. (2005) Mol. Cell. Biol. 25, 1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. (2007) J. Biol. Chem. 282, 7641–7655 [DOI] [PubMed] [Google Scholar]
- 26.Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P. (2006) Mol. Cell 23, 109–119 [DOI] [PubMed] [Google Scholar]
- 27.Purbey P. K., Jayakumar P. C., Patole M. S., Galande S. (2006) Nat. Protoc. 1, 1820–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Driscoll R., Hudson A., Jackson S. P. (2007) Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. (2007) Science 315, 653–655 [DOI] [PubMed] [Google Scholar]
- 30.Tang Y., Holbert M. A., Wurtele H., Meeth K., Rocha W., Gharib M., Jiang E., Thibault P., Verrault A., Cole P. A., Marmorstein R. (2008) Nat. Struct. Mol. Biol. 15, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 33.Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tjeertes J. V., Miller K. M., Jackson S. P. (2009) EMBO J. 28, 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan J., Pu M., Zhang Z., Lou Z. (2009) Cell Cycle 8, 1747–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyrylenko S., Kyrylenko O., Suuronen T., Salminen A. (2003) Cell Mol. Life Sci. 60, 1990–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celic I., Masumoto H., Griffith W. P., Meluh P., Cotter R. J., Boeke J. D., Verreault A. (2006) Curr. Biol. 16, 1280–1289 [DOI] [PubMed] [Google Scholar]
- 38.Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. (2008) Cell 134, 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dryden S. C., Nahhas F. A., Nowak J. E., Goustin A. S., Tainsky M. A. (2003) Mol. Cell. Biol. 23, 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaquero A., Scher M. B., Lee D. H., Sutton A., Cheng H. L., Alt F. W., Serrano L., Sternglanz R., Reinberg D. (2006) Genes Dev. 20, 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scher M. B., Vaquero A., Reinberg D. (2007) Genes. Dev. 21, 920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., Ørntoft T., Lukas J., Bartek J. (2005) Nature 434, 864–870 [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Capetillo O., Allis C. D., Nussenzweig A. (2004) J. Exp. Med. 199, 1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.