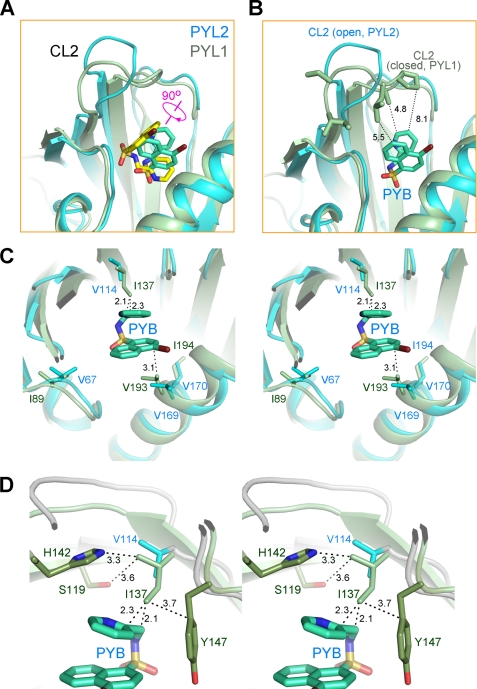
FIGURE 3.
Pyrabactin adopts distinct orientations in PYL1 and PYL2. A, pyrabactin recognition by PYL1 and PYL2 is compared. The structures of pyrabactin-bound PYL1 and PYL2 protomers are superimposed. PYL1 protein is shown in pale green, with the bound pyrabactin shown as yellow sticks. PYL2 is shown in cyan, with the bound pyrabactin shown as cyan sticks. Note that the bound pyrabactin is rotated for ∼90 degrees in PYL2 relative to that in PYL1. B, orientation of pyrabactin in PYL2 cannot stabilize a closed conformation of CL2. Structures of pyrabactin-bound PYL1 (pale green) and PYL2 (cyan) are superimposed as in A, and the pyrabactin in PYL2 is shown as cyan sticks. CL2 is unlikely to form enough contacts with pyrabactin in PYL2 even if it is closed as in PYL1. C, in PYL1, the side chain of Ile137 is too close to pyrabactin if the ligand were localized as in PYL2. Structures of pyrabactin-bound PYL1 and PYL2 are superimposed as in A. The pyrabactin in PYL2 is shown as cyan sticks. D, surrounding residues may prevent Ile137 from adopting other rotamers. Rotation of the side chain of Ile137 for 90, 180, or 270 degrees may cause potential clash with the side chains of Ser119, His142, and Tyr147.