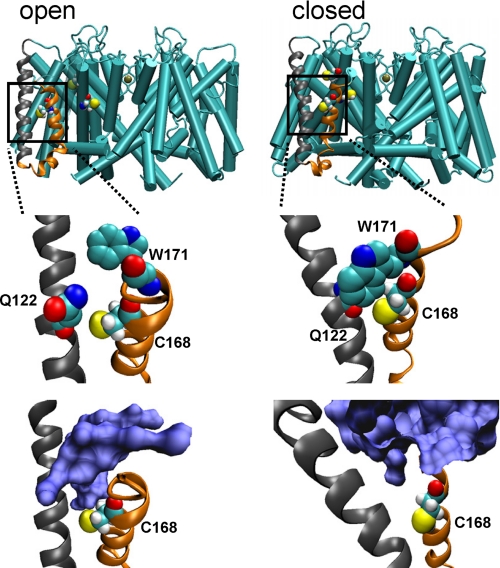
FIGURE 6.
Cys168 residue of SKOR is expected to reside in a water-filled pocket accessible from the outside in the open, but not in the closed conformation. Results of molecular dynamic simulations are shown in a side-on view of the SKOR channel assembly (top) with S2 and S3 α-helices of one monomer in ribbon (gray and orange, respectively) and Cys168, Cys228, and Cys234 in van der Waals representations following equilibration (see supplemental Fig. S1). Expanded views of the S2 and S3 α-helices show amino acid residues (center) and the water-filled space in blue (bottom) adjacent to Cys168. Protein structures were recorded every 10 ps and the root mean square deviation was calculated with the VMD trajectory tool (46) using α-C atoms to confirm equilibration. Root mean square deviation averages from 300 superimposed coordinates were determined at 10-ps intervals and gave a root mean square deviation of 0.863 ± 0.083 (±S.D.) in the open state and 0.457 ± 0.035 (±S.D.) in the closed state.