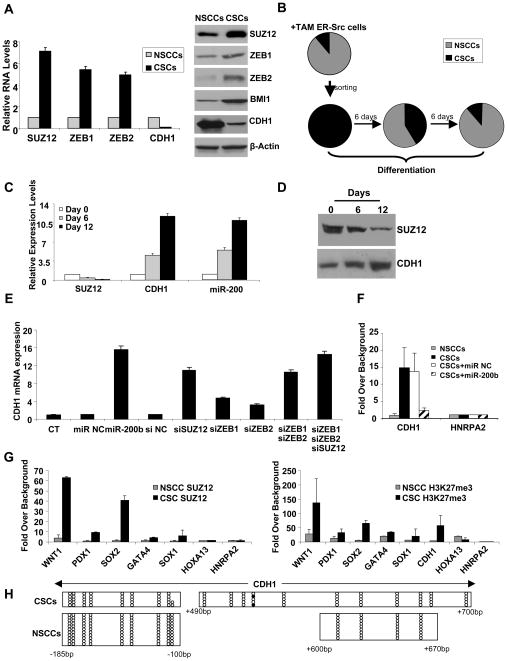
Figure 4. MiR-200b Functions through Suz12 and H3K27 Tri-methylation in CSCs.
(A) Suz12 and E-cadherin (Cdh1) mRNA (left) and protein (right) levels (mean ± SD) in NSCCs and CSCs.
(B) Percentage (mean ± SD) of CSCs (black) and NSCCs (gray) at the indicated times after plating CSCs (obtained from sorting ER-Src transformed cells) under differentiation conditions.
(C) Suz12, Cdh1, and miR-200 RNA levels (mean ± SD) during differentiation of CSCs.
(D) Suz12 and Cdh1 protein levels during differentiation of CSCs.
(E) Cdh1 RNA levels (mean ± SD) in CSCs after transfection of miR-200b or siRNAs against the indicated genes.
(F) Chromatin immunoprecipitation showing association of Suz12 at CDH1 and control HNRPA2 gene in NSCCs and CSCs expressing miR-200 or a control microRNA. Data are presented as mean ± SD.
(G) Suz12 association and H3-K27 trimethylation (mean ± SD) at the indicated loci in NSCCs and CSCs.
(H) DNA methylation analysis of regions within the CDH1 promoter in CSCs and NSCCs. For each horizontal line, open circles indicate non-methylated residues and black circles indicated methylated residues in a given clone that was sequenced after bisulfite conversion.