Figure 7.
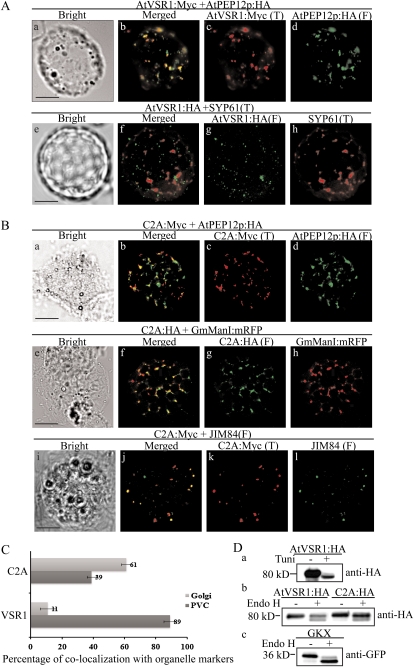
The majority of C2A:HA localizes to the TGN, with a minor proportion to the PVC. A and B, Localization of AtVSR1:Myc and C2A:Myc. Protoplasts were transformed with the indicated constructs, and localization of proteins was examined by immunostaining with the appropriate antibodies. The fluorescent signal of GmManI:mRFP was observed directly. F, Fluorescein isothiocyanate; T, tetramethylrhodamine-5-(and-6)-isothiocyanate. Bars = 20 μm. C, Quantification of overlap between C2A and organellar markers. The degree of overlap between C2A:Myc and JIM84 or AtPEP12p:HA was quantified by analyzing more than 50 protoplasts randomly selected from a population of transformed protoplasts each time. Three independent transformation experiments were performed. As a control, the degree of overlap between wild-type AtVSR1:Myc and organellar markers was quantified. Error bars indicate sd (n = 3). D, Endo H-resistant N-glycans of C2A:HA. Protoplasts transformed with the indicated constructs were incubated with or without tunicamycin (100 μg mL−1) for 24 h, after which protein extracts were prepared from transformed protoplasts and treated with or without endo H. GKX was included as a control for ER proteins. The samples were analyzed by western blot using anti-HA or anti-GFP antibodies.