Table 3.
Gene Top 10
| CBP T30 Versus T0 | p300 T30 Versus T0 | ||||
|---|---|---|---|---|---|
| Gene | P-value | Ratio | Gene | P-value | Ratio |
| CATSPER3 | 0 | 2.58 | THBS1 | 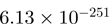 |
6.07 |
| ATF3 | 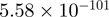 |
4.51 | ATF3 | 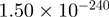 |
5.83 |
| TRIP13 | 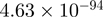 |
11.07 | FOSB | 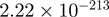 |
14.03 |
| CYR61 | 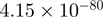 |
6.43 | CYR61 | 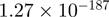 |
6.33 |
| FOSB | 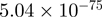 |
10.03 | EGR1 | 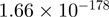 |
14.11 |
| SMAD3 | 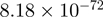 |
2.09 | TPM1 | 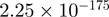 |
4.81 |
| TMEM49 | 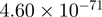 |
2.32 | DUSP1 | 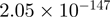 |
6.81 |
| MYH9 | 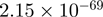 |
2.39 | MYH9 | 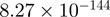 |
2.84 |
| CRISPLD2 | 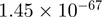 |
2.89 | NR4A1 | 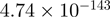 |
13.16 |
| THBS1 | 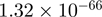 |
4.15 | CRISPLD2 | 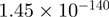 |
4.04 |
| T0 p300 Versus CBP | T30 p300 Versus CBP | ||||
|---|---|---|---|---|---|
| Gene | P-value | Ratio | Gene | P-value | Ratio |
| CXXC1 | 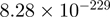 |
22.71 | CXXC1 | 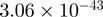 |
20.32 |
| AKT1S1 | 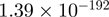 |
6.03 | MKKS | 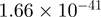 |
3.87 |
| FBXL19 | 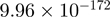 |
5.56 | CATSPER3 | 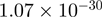 |
1.44 |
| MKKS | 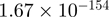 |
4.73 | AKT1S1 | 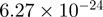 |
4.46 |
| C3orf19 | 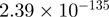 |
7.93 | FAM40A | 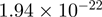 |
4.21 |
| BSCL2 | 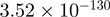 |
8.25 | FBXL19 | 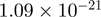 |
3.27 |
| THBS1 | 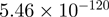 |
2.1 | ZNF350 | 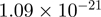 |
9.14 |
| MADCAM1 | 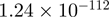 |
7.85 | METTL3 | 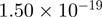 |
13.47 |
| ZNF175 | 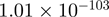 |
17.86 | MADCAM1 | 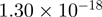 |
7.2 |
| C1orf174 | 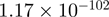 |
9.84 | C1orf174 | 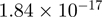 |
7.25 |
Top 10 genes that are most significantly different between time points and coactivators according to the P-values of the Fisher’s exact test. The ratio shows the quantitative difference in binding as expressed by the number of tags between the two samples that are compared: from T30 and T0 (upper half of the table) and from p300 and CBP (lower half of the table).
