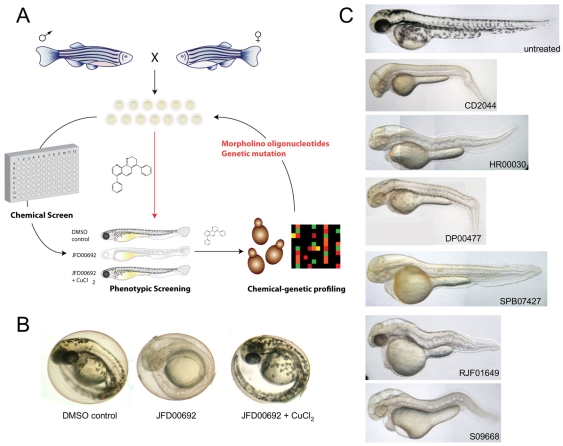
Fig. 1.
A combined zebrafish and yeast approach to probe chemical-genetic interactions in melanocyte pigmentation. (A) Overview of work flow. Phenotypic screening in zebrafish informs as to the developmental and physiological target pathway. Small molecules that promote similar phenotypes in zebrafish are then tested in genome-wide chemical-genetic screens in yeast to reveal potential target genetic pathways. Conserved target pathways are then directly re-tested for chemical-genetic interactions in the zebrafish system using genetic mutant lines, morpholino oligonucleotides or small molecules known to impinge on the same target. (B) Phenotypic screening of a >1500-compound yeast-bioactive library (S.D., J.W., M. Spitzer and M.T., unpublished data) and the Sigma LOPAC library for altered pigmentation in zebrafish embryos revealed a copper-metabolism phenotype characterized by hypopigmentation, an expanded hindbrain and an undulating notochord (Mendelsohn et al., 2006). Representative phenotype caused by 10 μM JFD00692 was rescued by treatment with exogenous copper chloride (5 μM or 15 μM) during development. (C) Examples of the copper-metabolism phenotype induced by some of the compounds identified in the screen.