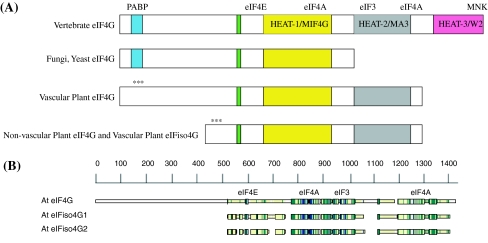
Fig. 1.
a eIF4G and eIFiso4G Domain Organization. The organization of eIF4G from vertebrates, fungi/yeast and plants have in common the eIF4E binding domain and core HEAT domain for binding eIF4A and eIF3 (MIF4G). Fungi and yeast have lost the second and third HEAT domains that include the second eIF4A binding domain and MNK binding domain (Hernandez and Vazquez-Pianzola 2005; Marintchev and Wagner 2005; Schutz et al. 2008). Plant eIF4G and eIFiso4G have retained only the first and second HEAT domains. The PABP binding domain for plant eIF4G or eIFiso4G (***) has not been experimentally determined. b Amino Acid Sequence Comparison. The amino acid sequences for Arabidopsis thaliana eIF4G (At3g60240) and the two eIFiso4G (At5g57870, At2g24050) proteins were aligned using CLUSTALW2 (Larkin et al. 2007) and visualized with MACAW (v.2.0.5) (Schuler et al. 1991). The approximate binding domains for eIF4E/iso4E, eIF4A and eIF3 are indicated. The actual sequence alignments are shown in Supplemental Data Figure B. The PABP binding domain (***) for plant eIF4G or eIFiso4G has not been experimentally determined