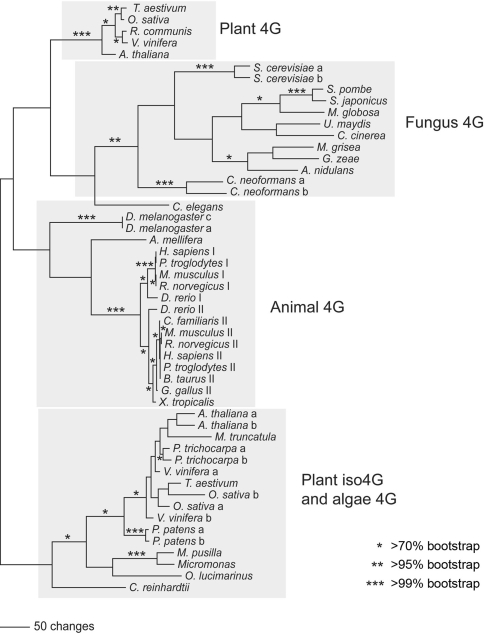
Fig. 2.
eIF4G/eIFiso4G Phylogeny. Amino acid sequences from the conserved HEAT-1/MIF4G domain were aligned for 50 eIF4G or eIFiso4G sequences from plants (vascular and non-vascular), animals, and fungi using the program MAFFT (Katoh and Toh 2008). A phylogenetic analysis of these aligned sequences (see Supplementary data Fig. C) was then conducted using PAUP* (Swofford 2000), under the parsimony, minimum evolution, and maximum likelihood criteria (the parsimony results are shown here), with bootstrapping used to assess support. Asterisks indicate >70% (*), >95% (**), and >99% (***) bootstrap support