Abstract
Artificial chromosomes and minichromosome-like episomes are large DNA molecules capable of containing whole genomic loci, and be maintained as nonintegrating, replicating molecules in proliferating human somatic cells. Authentic human artificial chromosomes are very difficult to engineer because of the difficulties associated with centromere structure, so they are not widely used for gene-therapy applications. However, OriP/EBNA1-based episomes, which they lack true centromeres, can be maintained stably in dividing cells as they bind to mitotic chromosomes and segregate into daughter cells. These episomes are more easily engineered than true human artificial chromosomes and can carry entire genes along with all their regulatory sequences. Thus, these constructs may facilitate the long-term persistence and physiological regulation of the expression of therapeutic genes, which is crucial for some gene therapy applications. In particular, they are promising vectors for gene therapy in inherited diseases that are caused by recessive mutations, for example haemophilia A and Friedreich's ataxia. Interestingly, the episome carrying the frataxin gene (deficient in Friedreich's ataxia) has been demonstrated to rescue the susceptibility to oxidative stress which is typical of fibroblasts from Friedreich's ataxia patients. This provides evidence of their potential to treat genetic diseases linked to recessive mutations through gene therapy.
1. The Rationale behind the Use of Large DNA Molecules in Gene Therapy
Gene therapy is defined as the transfer of nucleic acid molecules (usually DNA) to a patients somatic cells in order to prevent, treat, or alleviate a specific condition. Different gene therapy strategies have been designed to suit different types of diseases, the most “classical” of which involves gene delivery to target cells in order to obtain optimal expression of the gene introduced. This therapeutic approach is particularly well suited for inherited diseases that are caused by recessive mutations, since these are typically associated with the absence of a functional gene product or the drastic decrease in the expression of a gene. In these cases, the “therapeutic gene” must be inserted within a DNA molecule (usually a bacterial plasmid) along with all its essential regulatory sequences in order to ensure the correct expression of the gene in the target cells. To facilitate the adequate cellular uptake of DNA molecules, they must be packed within appropriate “gene delivery vehicles”.
Viral vectors have become the preferred “gene delivery vehicles” in the field of gene therapy due to their extremely high efficiency of gene transfer in somatic cells. However, viral vectors fail to fulfil all the requirements for an “optimal” gene therapy vector. Ideally, the best gene therapy protocol would involve a single administration of the therapeutic gene to the organism, such that it could replicate and segregate alongside the endogenous chromosomes, if necessary, thereby permitting long-term maintenance in any dividing cell. However, most viral vectors are either nonintegrating episomes that are unable to replicate, segregate, and persist in dividing cells, or they are integrating vectors. These latter vectors insert into the chromosomes of the host cell, and although they can therefore persist in proliferating cells, they carry the risk of producing insertional mutagenesis.
Adenoviruses are DNA viruses that have a linear double-stranded 36 kb genome. These viruses can be used as nonintegrating vectors since their genome is maintained as an episome once it is released into the nucleus of the host cell. However, these episomes are lost after several cell divisions. Moreover, the first-generation adenoviral vectors elicited a strong immune response that led to elimination of the transduced cells, and the humoral response excluded them after future administrations [1, 2]. To diminish or overcome this reaction, multiple deletions have been introduced into the subsequent generations of high-capacity [3] “gutless” adenoviral vectors [4, 5]. Nevertheless, while these vectors can now infect a wide range of cells, their lack of persistence in dividing cells represents an important weakness.
Retroviral-based vectors are capable of transducing a wide variety of dividing cells and they are efficiently integrated in the genome of the host cells [6]. However, this latter characteristic represents their major disadvantage since random integration may cause insertional mutagenesis, and the ensuing activation and/or silencing of undesirable genes might even trigger oncogenic transformation in the host cells [7, 8]. Indeed, some patients treated with retroviral vectors in a clinical trial for severe combined immunodeficiency (SCID) developed leukemia as a consequence of vector integration [9].
While Lentiviruses belong to the retrovirus family, they are able to transduce both proliferating and non-proliferating cells, augmenting the range of target cells [10, 11]. These viruses are currently the most widely used gene therapy vectors, as they sometimes permit prolonged gene expression, although gradually silencing of gene expression has also been reported on occasion [12].
Finally, most viral vectors have a limited packaging capacity and they cannot accommodate large genes. For this reason, most constructs used for gene therapy are “minigenes”, consisting of cDNA sequences (instead of entire genes) under the control of heterologous promoters (usually small compacted promoter sequences of viral origin). These “minigene constructs” often exhibit variable expression or lack tissue specificity, and they may not be properly regulated in somatic cells, being completely “silenced” in many cases, which obviously restricts their usefulness in gene therapy. Indeed, studies performed in transgenic mice have demonstrated that these “minigene constructs” are very susceptible to being switched off by neighbouring chromatin [13]. On the other hand, there are cases where the cDNA is too big to be contained in one of these vector systems and some “nonessential” regions have to be eliminated [14].
By contrast, studies performed in transgenic mice have highlighted the advantages of using large fragments of genomic DNA that permit the delivery of intact mammalian genes with all their introns, promoters, enhancers, and long-range controlling elements. By using gene's normal promoter and controlling elements, the level and control of expression is comparable to the endogenous expression of the gene. The experience gained from using yeast artificial chromosomes (YACs) and bacterial artificial chromosomes (BACs) has shown that large genomic fragments drive tissue-specific expression at endogenous levels [15–20]. For example, transgenic mice carrying the intact human cystic fibrosis transmembrane regulator gene (CFTR), which spans 200 kb of a 300 kb YAC [21], express CFTR in an appropriate tissue specific manner and complement the cftr defect in null mice. Similar physiological expression has also been achieved with the Huntingtin gene, which has been difficult to express from minigene constructs [22]. Likewise, the use of a YAC or BAC carrying the entire frataxin gene has rescued frataxin knock-out mice from embryonic lethality [16, 19]. In view of these data, large DNA molecules seem to be better candidates as vectors for gene therapy than the typical viral vectors, with their limited capacity.
As a result of the human genome project, BACs are available that cover the whole genome. Given that the average size of a BAC is approximately 150 kb, most genes, together with all their regulatory elements, can be included in such a construction. A system for linking overlapping BACs by homologous recombination in E. coli has been described for when a BAC containing a whole locus is not available, or if new regions need to be added upstream or downstream of a given gene within a BAC [23–25]. Also recombination has been used to modify BACs with different tags, which is very useful in studies of protein-protein or DNA-protein interactions, and of protein localization [26].
This paper will deal with mammalian artificial chromosomes and minichromosome-like episomes. We will focus on recent advances on the use of BACs containing genomic loci as a platform for the design of artificial minichromosome-like vectors for gene therapy applications, and on the “delivery vehicles” that can accommodate such large DNA molecules. We will also analyse two examples of applications of these artificial minichromosome-like vectors for rare genetic diseases (haemophilia A and Friedreich's ataxia).
2. Mammalian Artificial Chromosomes
An obvious vector to stably carry large genomic DNA fragments would be an artificial minichromosome that must have a replication origin and a centromere, enabling the DNA molecule to replicate during S phase and to correctly segregate during mitosis. In addition, if the DNA molecule is lineal telomeres must be present at both ends to guarantee its stability. The centromere is the most important chromosomal element since it is crucial for the attachment of chromosomes to the mitotic spindle and their correct segregation in mitosis. Centromeric DNA binds the CENP proteins that form the scaffold for the kinetochore, the structure connecting the centromere to the spindle microtubules. Artificial chromosomes have been built in yeast without any difficulty, since yeast chromosomes contain small centromeres. However, mammalian centromeres are very large and extraordinarily difficult to engineer, which has complicated the construction of mammalian artificial chromosomes (MACs) or human artificial chromosomes (HACs) [27].
2.1. Chromosomal Elements
All normal human centromeres contain large arrays of alphoid satellite DNA (α-DNA), a family of repetitive DNA sequences first discovered in the African green monkey [28]. Satellite DNA corresponds to only a small fraction of the human genome (less than 2%) whereas in other species like the rat, it is much more abundant [29]. Human α-DNA consists of tandem repeats of a 171 bp monomer arranged in highly ordered repeats [30, 31]. The divergent intermonomeric sequences lying between the independent monomers vary from 20% to 40% whereas the sequence divergence is around 5% in the higher order repeats [32]. Despite this monomeric variation, each human centromere is thought to be formed by arrays of a unique monomer spanning from several kilobases up to megabases [33, 34].
Several attempts to generate artificial chromosomes have used alphoid DNA arrays from different chromosomes (X, Y, 17, and 21), although not all have succeeded. Alphoid DNA from chromosome Y has failed to form active centromeres in some cases [35, 36], probably because of the lack of CENP-B boxes [37]. Masumoto and collaborators have also showed how two different alphoid DNA loci from the same chromosome behave in a completely opposite manner. They found that alphoid-derived YAC unable to form HACs do not have CENP-B boxes in addition to a more diverged alphoid repeats [38].
In terms of specific DNA sequences, alphoid DNA may not be the only element needed for centromeres to fulfil their function, implicating additional mechanisms [39]. This view is supported by the lack of a consensus sequence for α-DNA [32], the existence of dicentric chromosomes with two identical regions of α-DNA but only one active centromere [40–42], or the fact that functional neocentromeres can be formed from noncentromeric DNA [43]. However, it is clear that no DNA other than alphoid DNA can efficiently form a centromere after transfection into human cells. It has been already mention that not all alphoid DNAs can form active centromeres with the same efficiency and it appears that functional centromeres required a minimum size, estimated to be around 100 kb [44]. Lo and collaborators analysed all chromosomes in order to determine the minimum length of alphoid DNA tolerated by chromosomes and still able to seed de novo centromere formation [45]. In general, chromosomes cannot bear an important reduction in the length of alphoid DNA array, with the exception of chromosome 21 whose alphoid DNA could oscillate between 50 and 100 kb, with an average size of 78 kb.
Despite all this evidence, there are several reports where introducing different arrays of alphoid DNA does indeed lead to centromere formation. Several studies have shown how when large (>50 kb) arrays of alphoid DNA are transfected into a human cell line (HT1080) in culture, de novo chromosomes may form with a functional centromere [35, 46, 47]. Nevertheless, much work is still needed in order to elucidate the mechanisms underlying de novo centromere formation.
2.2. MAC Formation
There are two different strategies to form “de novo” HACs. The “bottom-up” approach consists of assembling the elements needed for HAC formation in a vector and with this aim, a universal system to introduce alphoid DNA into any BAC [48] using in vivo recombineering has been design, containing a 70 kb array of this alphoid DNA proven to be capable of forming de novo MACs [38, 46]. With the second strategy or “top-down” approach an existing chromosome is manipulated in order to reduce its size, eliminating unneeded elements except for the centromere region [49].
In the first studies carried out on the formation of centromeres and de novo human artificial chromosomes (HACs) [35], synthetic alpha satellite arrays were cotransfected with telomeric DNA and other genomic sequences to obtain linear minichromosomes of 6 to 10 Mb in size. Due to the already mention sequence divergence between alphoid DNA, arrays from 2 different chromosomes were used to obtain 4 minichromosomes-containing cell lines, two with alphoid DNA from the Y chromosome and the other two with alphoid DNA from chromosome 17. However, three of these minichromosomes had acquired DNA from other centromeres or neo-centromeres, and only one of them contained alphoid DNA exclusively from the transfected α-DNA. The HACs were cytogenetically stable, in terms of the presence and absence of selection, and their copy number or size did not change over time.
Only one year later, YAC-based mammalian artificial chromosomes (MACs) were generated [47], yeast artificial chromosomes having been widely used to introduce mammalian genes into mice as they can harbour large DNA fragments [15, 50, 51]. In these studies, a recombination deficient yeast host was used in which alphoid arrays from chromosome 21 (α21-I and α21-II), telomere sequences and selectable markers were introduced, and the modified YAC was transfected into HT1080 cells. The results obtained showed some variability regarding the proportion of cell lines containing minichromosomes (from 18% to 68%), although the number of MACs per cell line was most frequently one. The MACs obtained segregated accurately, they bound centromeric proteins, and they were about 1–5 Mb in size. In addition, they did not gain host DNA and presented a loss rate of less than 1% per generation in the absence of selection.
A further advance came when it was demonstrated that no telomere sequences are needed when the input DNA is circular [46]. Accordingly, it was established that MACs can be formed in HT1080 cells from either linear or circular input DNA, with circular alphoid constructs being very effective at forming MACs, whether or not human telomere sequences are added. Indeed, the presence of these arrays did not favour MAC formation. By contrast, it was ascertained how linear DNA molecules require telomere sequences to maintain and protect minichromosome ends.
When considering the de novo formation of HACs, an important inconvenience is the fact that these structures are not stable in murine cell lines [52], making it difficult to perform studies in mice as a model system. When a previously established HAC was transferred into several murine cell lines (RAG, STO and LA-9), the HAC DNA rearranged and was different to the original HAC. However, murine DNA was not acquired and no integration events were observed. When selection was withdrawn, the artificial chromosomes were rapidly lost at a rate between 2%–5% per day, depending on the cell line. These results show how centromere activity is retained to some extent, as they were still able to bind a marker of active centromere, yet as pointed out above, they also suggested that something else is needed to provide correct segregation.
2.3. Gene Expression from Artificial Chromosomes
The next advance in the development of human artificial chromosomes was probably the demonstration that any gene cloned into those constructs could be expressed normally, as addressed in several reports. There are two studies where the entire human HPRT gene was expressed from such mammalian artificial chromosomes (MACs) [53, 54], using slightly different strategies but with the same results. A PAC containing about 140 kb of genomic DNA, including the human HPRT gene, and a second PAC including a 70 kb array of α-DNA derived from chromosome 21 (α21-I) [46] were transfected into HPRT-deficient HT1080 cells [53]. The results obtained showed how two of the three MAC-containing cell lines generated were mitotically stable and that HPRT expression was maintained over long periods of time, even in the absence of selection. In a second study, a 404 kb HAC made by in vivo recombination and introduced into HPRT-deficient HT1080 cells successfully ensured expression of the gene after two months without any selection [55].
More recently, a 21-derived minichromosome made in DT-40 chicken cells using the top-down approach [56] was introduced into human HT1080 cells by microcell-mediated chromosome transfer to check its stability [57]. Subsequently, the erythropoietin gene was added by cre-loxP recombination and transgene expression and vector stability was finally tested in HFL-1 cells. Accordingly, the vector was mainly maintained as a single copy per cell with no translocation and/or insertion events, and epo expression was sustained over 12 weeks without any loss in the absence of selection.
Using the same strategy, the dystrophin gene (about 2.4 Mb in length) has been cloned into a HAC [58], a construction that enabled all the tissue-specific isoforms of human dystrophin gene to be detected, overcoming the problem shared by other vector systems that only produce one of the isoforms. One year later, the same group used the dystrophin HAC to correct patient-derived fibroblasts and then transform the corrected cells into human iPS (induced pluripotent stem) cells [59]. This is a great achievement as probably autologous-corrected cells would be the best candidate cells to be used in gene therapy protocols.
2.4. Conclusions
It seems that the size and/or structure of the HAC matters since artificial chromosomes have a higher rate of missegregation than normal chromosomes [60, 61]. It is also notable that different HACs behave in a different way in different cell lines [62], suggesting that centromere activity could probably be maintained by trans-acting factors with different specificities between different cell lines, even within the same species.
A better understanding of centromere function seems crucial in order to be able to generate “synthetic” centromeres, which should improve the development of MACs. Thus, despite some of the advances and successes, the engineering of HACs/MACs to carry genes of interest is evidently quite difficult, which has restricted their use as vectors for gene therapy.
3. Stable Replicating Episomes
In view of the difficulties associated with MACs, other alternative vectors have been developed to carry large genomic constructs. Among them, stable non-integrating and replicating episomes have emerged as very promising gene therapy vectors for dividing cells. Interestingly, these stable episomes do not become integrated within host cell chromosomes and are therefore not associated with any risk of insertional mutagenesis.
3.1. OriP/EBNA-1 Vectors: Episomal Maintenance
Perhaps the best characterized artificial minichromosome-like episomes of this type are the so called OriP/EBNA1 vectors. These vectors are bacterial plasmids or bacterial artificial chromosomes in which episomal replication and segregation are achieved through DNA sequences derived from the Epstein-Barr virus (EBV). This herpes virus has a genome consisting of dsDNA of approximately 172 kb and it is characterized by a latent phase during which it is maintained for life asymptomatically in lymphocytes of about 90% of the human population. In the latent stage, the viral genome is maintained as a circular extrachromosomal replicating episome within the nucleus of infected cells [63].
The only viral elements needed to confer replication and segregation on this episome are the latent origin of replication, oriP, and the viral gene encoding the EBNA1 protein [64–66]. The origin of plasmid replication (oriP) from Epstein-Barr virus is a cis-acting element which has been proven to confer replication autonomy and maintenance to recombinant plasmids in cells harbouring latent EBV. OriP is formed by two elements about 1 kb apart: the family of repeats (FR), a 20 member family of 30 bp direct repeats, and the dyad symmetry (DS) element, a 65 bp DS containing four copies of the repeat [67, 68]. OriP resides within a 1.8 kb segment in the short unique region of the EBV genome, and it appears to be the only element needed in cis [65, 68]. The trans-acting gene allowing oriP function lies in a 2.6 kb region encoding the Epstein-Barr Nuclear Antigen 1 (EBNA-1) [66]. The EBNA-1 protein can bind to both the FR and DS sequences [69], whereby the DS element is the initiation site of episomal DNA replication whereas FR acts as a replication fork barrier and termination site [70]. EBNA-1 also activates the 30 bp repeats of oriP in trans, the latter acting as a transcriptional enhancer for genes linked to the repeat [71].
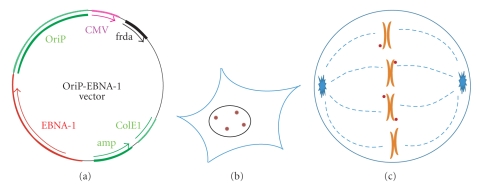
Plasmids containing these two elements (oriP and EBNA-1) are replicated once per cell cycle and they segregate passively by attachment to the mitotic chromosomes [72, 73]. Such plasmids are maintained in human cell lines in tissue culture [63, 66], as well as in mouse cell lines if the plasmid contains large fragments of genomic DNA that permit replication [74]. Although standard EBV-derived vectors usually carry both oriP and EBNA-1, it is well established that vectors containing only the oriP FR and EBNA-1 as well as inserts of genomic DNA greater than 100 kb can be replicated and maintained as episomes, and can thus be used for studies in human and rodent cells [63]. As the segregation of episomes is passive, by binding to endogeneous chromosomes instead of by direct attachment to the microtubules of the mitotic spindle, selection is sometimes required to maintain a population of cells that carry the episomes (especially in cases of a low copy number of episomes per cell). The typical structure of OriP/EBNA1 vectors and their localization in the interphase and mitotic cells is shown in Figure 1.
Figure 1.
(a) Structure of oriP/EBNA-1 vectors. The scheme represents a frataxin expression plasmid-containing episomal elements (oriP/EBNA-1). (b) orP/EBNA-1 plasmids remain as stable replicating, nonintegrating episomes within nuclei of interphase (and postmitotic) cells. (c) oriP/EBNA-1 episomes attach to the periphery of chromosomes through EBNA-1 protein and segregate during mitosis.
The replication and segregation of OriP/EBNA1 vectors depends only on the presence of EBNA-1 protein. As mentioned above, the binding of EBNA-1 to the DS element of OriP destabilizes nucleosomes and aids the recruitment of cellular replication factors to OriP. The segregation function of EBNA-1 depends on its ability to bind to both the FR element of OriP and to the periphery of chromosomes. It is thought that EBNA-1 interacts with chromosomes in two different ways. During interphase (and probably also in mitosis), EBNA-1 associates with chromatin through its binding to either cellular DNA or chromatin-associated proteins. During metaphase/anaphase, there is additional binding of EBNA-1 to a cellular protein referred to as EBP2 (EBNA1-binding protein 2), which strengthens the association of EBNA-1 with mitotic chromosomes [75].
3.2. Applications
OriP/EBNA-1-based vectors have been widely used for a variety of purposes. One of the strategies for cloning a eukaryotic gene relies on the ability to select for the functional expression of the gene of interest in mammalian cells [76]. A subcloning vector has been designed containing oriP/EBNA-1 that enables the expression of very rare clones to be selected directly [77]. These clones were stably maintained as episomes, 2 to 10 copies per cell, as long as selection was applied.
Other studies using oriP/EBNA-1 vectors have expressed cDNA from heterologous promoters, all producing high levels of expression for several weeks or months, and all showing stable retention of the episomes in the cells for long periods of time [78–81]. Such vectors have even been used to produce large amounts of recombinant proteins [82].
There are also several studies showing how these two elements are sufficient for large plasmids to be maintained indefinitely in cells, expressing the gene they contain in a physiological fashion. For example, 2 YACs of 90 and 660 kb were created that contained oriP and when they were introduced into human cells expressing EBNA-1, these constructs were maintained as stable episomes for more than 8 months in the presence of selection, and for up to 5.5 months without selection [83]. Three cell lines were produced with the 90 kb YAC, all of which contained unrearranged episomes. However, of the 3 cell lines containing the 660 kb YAC only two presented the intact form of the 660 kb molecule. FISH analysis also demonstrated how episomes, some visible as pairs, associated with the host cell chromosomes. This association may explain how they are so efficiently maintained, even in the absence of selection, indicating that stability is achieved by attachment to host chromosomes as proposed for the EBV genome [84]. An intact CFTR gene has also been expressed from an oriP/EBNA1 episome in mouse cells [85]. By introducing circularized YACs into CMT-93 and LA-9 cells by fusion with yeast spheroplasts, nonrearranged YACs were obtained and maintained as episomes of either 320 or 640 kb. The copy number varied between 2 to 56 in the different cell lines obtained and the rate of loss varied from 0.4% to 5% in the absence of selection, very similar to the data from the 293-cell lines expressing oriP-YACs [83]. The level of expression of the exogenous CFTR gene was dependent on its copy number and each copy of the YAC produced about 20% of the level of each endogenous gene. A further advance was made by changing the constructs based on YACs to others based on BACs, as the latter are easier to use and manipulate, allowing purification of much larger quantities of DNA.
In order to modify BACs for their use in gene therapy protocols, an efficient system for retrofitting BACs with oriP/EBNA1 and reporter genes has been described using loxP/Cre recombination [86, 87]. These vectors not only contain the elements needed for episomal maintenance, they also include suitable marker genes for selection and monitoring of the mammalian cells into which the constructs are introduced.
4. Delivery Methods
As mentioned above, the type of DNA molecules used for gene therapy is crucial but the “DNA delivery vehicles” are also very important. Indeed, the transfer of large DNA molecules to mammalian cells is not an easy task. Medium size BACs can be delivered into mammalian cells using Lipofectamine 2000, and more than half of the transfected cells contain intact delivered BAC [87, 88]. However, when working with BACs, the purification of supercoiled DNA may be quite cumbersome and it becomes increasingly difficult with larger BACs. Different methods involving less manipulation and/or higher efficiency of delivery should be tested in order to optimize the whole system. To date, two methods appear to be quite promising to transfer BACs-based vectors to mammalian cells: bactofection and packaging within Herpes simplex virus 1 (HSV-1) particles.
4.1. Bactofection
Bactofection refers to the use of bacteria for the delivery of DNA molecules to mammalian cells. One advantage of this method is that it minimizes the need to manipulate the DNA molecules, which is especially useful in the case of very large molecules such as BACs. Indeed, different studies have proven how DNA transfer can occur from bacteria to mammalian cells [89–91]. Hence, bacterial strains that are naturally or artificially engineered to be invasive, but that are attenuated to prevent pathogenesis, are particularly useful for bactofection. Accordingly, attenuated intracellular bacteria engineered to lyse after cell invasion have been shown to transfer functional genes to a very broad range of mammalian cells [92, 93]. The main bacterial species used as delivery vectors are facultative intracellular pathogens such as Salmonella sp [94], Shigella sp [95], Listeria sp [96], or Yersinia sp [97]. However, more recently, it was demonstrated that almost any species can be used provided they are appropriately engineered [98]. Anaerobic or facultative anaerobic bacteria are also being applied as anticancer agents to target solid tumours taking advantage of their ability to grow in the hypoxic region of tumours [96, 98–103]. Indeed, bacterial delivery has been assessed as a way to administer DNA vaccines using different intracellular pathogen strains [104, 105]. For certain purposes, determined bacterial toxins have been used as vectors to deliver DNA into mammalian cells [106].
A different approach for bacterial delivery relies on the use of a genetically modified E. coli strains that express two additional genes; inv from Yersinia pseudotuberculosis, and hly from Lysteria monocytogenes [107, 108]. Inv permits binding to the integrins expressed on the surface of mammalian cells and hly, the escape from lysosomes once in the cytosol of the mammalian cells. E. coli is also auxotrophic for aminopimelic acid (DAP−), which means that it is not able to form a new cellular membrane once in the mammalian cytosol. For this reason it lyses and freely liberates the DNA it contains. The Y. pseudotuberculosis invasin protein (inv) is encoded by a 3.8 kb gene expressing a 103 kDa protein localized to the outer membrane, part of a family of adhesins encoded by enteropathogenic bacteria [109]. All the members of this family have a region of similarity in the amino terminal 500 amino acids of the Y. pseudotuberculosis invasin (36% identity). This conserved domain is required for protein localization in the outer membrane and for export of the carboxyl termini of these proteins. Indeed, the terminal 192 amino acids of invasins represents the most divergent region and it is that which binds to integrin receptors [110, 111]. Invasin is the only protein needed to invade mammalian cells and it can be cloned into non-invasive strains of bacteria making them invasive [89, 112]. The second gene introduced in the modified E. coli strain is the hly locus from L. monocytogenes, encoding listeriolysin O. This protein is a member of the cytolysin family that are produced by various Gram positive species [113]. This 58 kDa cholesterol-dependent pore-forming toxin [114] allows the bacteria to escape from phagolysosomes into the cytosol of the infected cell [115]. In order to promote lysis of the internalized bacteria, the strain used is unable to synthesise a new cell wall when it divides since it is a diaminopimelic acid (dap) auxotrophic strain, a substrate not present in the mammalian cells. This ensures that the bacteria will not survive in the cells, and that the plasmid DNA will be liberated into the cytosol and eventually reach the nucleus.
Using this bacterial vehicle, functional DNA can be transferred into a variety of mammalian cell lines by simple coincubation with the engineered E. coli strain [107]. Different studies have showed that this strain is also able to transfer large DNA fragments [116–118], establishing the proof-of-principle for the use of this kind of delivery method as a true alternative to efficiently deliver intact BACs into recipient mammalian cells.
Currently, the use of bactofection seems to be limited to ex vivo gene transfer to cultured mammalian cells, although some examples of in vivo applications to cells of the gastrointestinal tract are envisaged. However, there are very serious concerns about the biosafety of bacteria as gene delivery vehicles due to the risks of important adverse immunological and inflammatory reactions, which will probably restrict their wider application in gene therapy.
4.2. Packaging into HSV-1 Virions
Vectors based on Herpes simplex virus 1 (HSV-1) represent one of the most promising alternatives for the delivery of large DNA molecules, including many artificial minichromosome-like episomes. HSV-1 has a genome of 152 kb, so viral vectors based on HSV-1 have a notably larger capacity to accommodate DNA than other conventional viral vectors used for gene therapy, such as retroviruses and adenoviruses [119].
HSV-1 derived amplicon vectors are plasmids (or BACs) bearing only two sequences of viral origin: an OriS to permit replication in packaging cells; and a pac sequence to permit packaging into HSV-1 viral particles. Hence, the remaining genomic capacity is available for exogenous DNA (up to approximately 150 kb), which may include entire genomic loci with all their regulatory elements to ensure persistent and physiologically regulated levels of expression [120–123]. This feature is probably the most promising characteristic of this type of vector as, to date, they are the only viral vectors with such a characteristic high efficiency of gene transduction that are capable of carrying such large DNA fragments that they may include a whole genomic locus. Other relevant aspects of HSV-1 amplicons as gene delivery vehicles include their capability to transduce both dividing and non-dividing cells, and their ability to persist as non-integrating episomes, eliminating the risk of insertional mutagenesis.
As amplicon vectors only contain the oriS and the pac packaging signal as viral DNA sequences, generating them requires the use of a helper virus encoding all the viral genes necessary for the assembly and packaging of DNA into viral particles. This helper virus, although replication-defective, quite often produces amplicons contaminated with helper viruses that can trigger immune and inflammatory responses [124]. Recently, a new system has been developed where all the genes needed for packaging are encoded in a large BAC (which is unable to be packed within virions), eliminating all the genes unnecessary to generate the amplicon-containing particles [123, 125, 126].
HSV-1 amplicons can play an important role in gene therapy protocols for neurological disorders due to their notable ability to deliver genes into neurons, both in vitro and in vivo [127–131]. Although they can express foreign genes driven by viral promoters only for a period of several days, genes under the control of a neuronal promoter can be stably expressed [132]. In this context, it is not surprising that expression from a genomic locus where the gene is under the control of its own promoter renders physiological levels of expression that are maintained stably [121]. As neurons are postmitotic cells, there is no need for reinoculation of the therapeutic gene should a gene therapy protocol use this kind of vector. However, for dividing cells some modifications would have to be made to the amplicon vectors in order for them to persist during cell divisions. One approach is to use hybrid amplicons containing elements from HSV-1 and elements from the Epstein-Barr virus that confer episomal retention [121, 122].
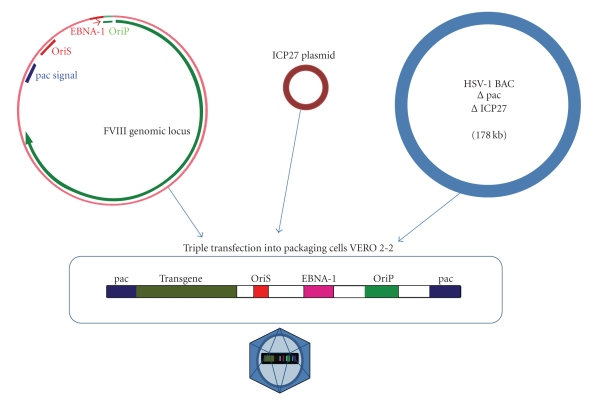
Figure 2 shows the structure of an HSV-1-derived amplicon vector encompassing an EBV-based minichromosome-like episome, and the procedure to pack it into HSV-1 virions. Given the high capacity of these vectors, even artificial minichromosomes may be delivered to mammalian cells with an efficiency higher than that obtained by other methods [133]. Thus, HSV-1 HAC amplicons containing alphoid DNA for episomal maintenance and a HPRT minigene have efficiently transduced cultured cells, producing stable HPRT expression for over three months. Curiously, not all HACs behave in the same way in the different cell lines tested, which was probably due to the level of expression of some proteins implicated in cell cycle control [133].
Figure 2.
Packaging of OriP/EBNA-1 episomes into herpesviral particles. HSV-1BAC and ICP27 plasmid are the elements used for packaging herpesvirus using the helper-free system.
Whereas HSV-1 virions are unable to pack DNA molecules larger than 160 kb, viral vectors based on other herpes viruses may have a greater capacity to accommodate exogenous DNA. Thus, cytomegalovirus, which has a 250 kb genome, may permit larger DNA fragments to be used [134]. In this way, herpes virus-based vectors appear to be highly promising “delivery vehicles” of artificial minichromosome-like episomes for gene therapy applications.
4.3. Gene Therapy of Haemophilia A
Haemophilia A is an X-linked recessive bleeding disorder caused by mutations in the Factor VIII gene (FVIII) that encodes for a clotting factor [135]. It affects about 1 in 5,000 male births in all populations and it is currently treated by infusion of plasma-derived or recombinant Factor VIII protein. The frequency of the disorder combined with the high cost of recombinant Factor VIII make it a major burden on the healthcare systems. Therefore, gene therapy for FVIII deficiency is clearly a very attractive option. There are different degrees of severity for this disease depending on the levels of the circulating-factors (severe, less than 1%; moderate, between 2%–5%; and mild, between 6%–30%), Thus, low levels of expression may have a large positive effect on the health of the individual. Interestingly, FVIII is normally expressed in the liver, an organ that is particularly easy to access for gene delivery. Much work has been carried out towards preparing gene therapies for both FVIII (haemophilia A) and Factor IX (FIX) deficiencies (hemophilia B). FIX is a much smaller protein encoded by a 1385 bp open reading frame that can be easily packaged as a minigene in retroviral, adenoviral and adeno-associated (AAV) vectors driven by either a viral or a mammalian promoter [136–139]. Long-term delivery and physiological expression have been achieved for FIX in mouse models, although many problems remain and gene therapy suitable for human patients is still not available [135]. By contrast, the open reading frame of FVIII is 7055 bp long and it has been much harder to express in viral vectors [140]. Different strategies have been applied to make the FVIII cDNA a bit shorter, and hence easier to introduce into different vectors, including using a B-domain deleted version of FVIII cDNA [141, 142] as this domain does not appear to be essential for coagulation [14, 143–145].
In view of these data, it seems that the election of the vector into which the FVIII gene has to be introduced is not a minor issue for haemophilia A gene therapy. First generation adenovirus-based vectors still contained viral gene sequences that produced immunogenic reactions and as a consequence, transgene expression was only short-lived as a result of vector clearance [146]. Nevertheless, even with these vectors it might be possible to achieve curative levels of FVIII for several months in haemophilic mice [147, 148]. However, the results from larger animals were not so promising [149, 150] and although some improvements in adenoviral vectors have been made, particularly regarding transgene expression, some toxicity still persists when these vectors are used [151, 152].
There is evidence of the expression of the FVIII cDNA with the B-domain deleted from retroviral vectors in different cells lines, from human skin fibroblasts [153, 154] to bone marrow stromal cells [155]. In each case, a functional FVIII factor is expressed but at low levels, due to the repressive sequences found in the cDNA of FVIII gene [156–159]. However, this difficulty has been overcome by using slightly different retroviral vectors based on the MFG retroviral vector system [160]. Yet, in these systems, the therapeutic levels of FVIII in circulation were obtained over just one week [161]. An important drawback of retroviral vectors is their inability to transduce nondividing cells, particularly important if gene therapy is directed to the liver. One way to bypass this limitation has been to use FVIII-expressing retroviral vectors to transduce neonatal mice where hepatocytes are undergoing rapid cell division, thereby obtaining complete correction of the disease [162]. A different approach is to employ lentivirus-based vectors which can transduce both dividing and non-dividing cells [163, 164].
Due to the small genome of adeno-associated virus (AAV) vectors, they have been used little with the FVIII gene. Nonetheless, there are reports of therapeutic levels of FVIII expression for a limited period of time using this viral system [165, 166]. Perhaps due to the disappointing results obtained with other viral vectors, delivery systems based on AAV vectors have recently attracted significant attention [167].
Besides the limitations presented by viral delivery systems, three phase I trials have been launched to test the safety of the procedure and to check the levels of expression from these vectors. In an ex vivo gene therapy protocol, a B-domain deleted FVIII cDNA expressed from a non-viral plasmid was electroporated into dermal fibroblasts and, after selection and expansion, the fibroblasts were reintroduced into the patients [168]. FVIII levels increased in four of the six subjects studied without producing adverse events. In the patient with the highest levels of FVIII expression, the therapeutic effects lasted for 10 months, although FVIII expression finally disappeared. In a second trial, a B-domain-deleted cDNA was expressed from a moloney murine leukaemia virus-based (retroviral) vector [169], which only produced levels of FVIII expression above 1% sporadically, concluding that efficient retroviral transduction would probably require higher doses and some degree of mitotic induction. In 2004, a third trial using an adenovirus vector encoding the complete FVIII cDNA was carried out in just a single patient. Despite obtaining sustained expression of FVIII above 1% of the normal levels for several months, adverse effects were observed associated with the appearance of thrombocytopenia and an elevation of transaminases [170].
There is only one study where factor VIII has been expressed from a BAC containing the entire genomic locus [118]. As no single FVIII-containing BAC was available, a new one was constructed by homologous recombination [171] and then retrofitted with different elements in order to provide episomal maintenance [48, 88]. The different vectors obtained were introduced into hepatic and nonhepatic human cell lines to check whether the construct could drive expression from the transgene, achieving detectable FVIII levels with all of them and in some cases, even stronger expression than the endogenous gene. Although a demonstration of functional FVIII expression from the BAC awaits the use of cell lines or mice with null endogenous expression, this is the first characterized genomic clone carrying the intact locus which could potentially be useful for therapeutic applications [118].
4.4. Gene Therapy for Friedreich's Ataxia
Friedreich's ataxia (FA) is the most common form of autosomal recessive ataxia and it is caused by a decrease in the levels of frataxin, a mitochondrial protein encoded by the nuclear FRDA (FXN) gene [172]. FA is a predominantly neurodegenerative disease with an estimated prevalence of 1-2 in 50,000 individuals, and it is characterized by the progressive loss of large sensory neurons and spinocerebellar tracts. Other clinical symptoms may include hypertrophic cardiomyopathy and diabetes mellitus.
The most common cause of the disease is an expansion of the GAA triplet within the first intron of the FRDA gene, which has a dramatic effect in reducing mRNA levels and consequently frataxin protein levels [173]. Although most patients have both chromosomes affected by the GAA expansion, it is possible to find some cases with one expanded allele and a point mutation in the other [174].
Frataxin is an 18 KDa protein [175] implicated in functions such as mitochondrial iron homeostasis [176, 177], iron-sulfur cluster biosynthesis [178] and oxidative phosphorylation [179]. Frataxin is expressed in all cell types, although cells from the nervous system, heart and muscle present the highest levels of the protein [180].
Homozygous frda knockout mice are embryonic lethal a few days after implantation, demonstrating the essential role for frataxin during early mammalian embryo development [181]. Thus, it appears that the milder phenotype associated with the human disease is due to the residual frataxin expression observed in patients with the expansion mutations. These results may also explain why no patients with homozygous point mutations have been encountered.
It has been difficult to generate mouse models for FA because of the embryonic lethality associated with the null mutations. Neuron-specific and conditional knock-out models generally exhibit a wider and more prominent neurodegenerative phenotype than the human disease [182, 183]. Some knock-in mice bearing the human gene with an expansion mutation surprisingly failed to develop any clinical phenotype, despite the significant reduction in frataxin levels to 25%–36% the wild-type levels [184]. More recently, representative FA mouse models have been generated by crossbreeding lines of a human FXN YAC transgenic mice that contain unstable GAA repeat expansions with heterozygous frda knock-out mice [185]. The resultant “transgenic-knockout” mice express comparatively little human-derived frataxin, and they exhibit a neurodegenerative and cardiac pathological phenotype similar to human disease sufferers, although significantly milder. With their limitations, these mice currently constitute the most useful model to study the physiopathology of FA and to test for possible therapeutic approaches.
There is no effective cure for FA, although antioxidants may have a mildly positive effect on some clinical signs [186]. There has been great interest in the possibility of boosting frataxin gene expression as a therapeutic approach and indeed, erythropoietin [187] has been shown to increase frataxin protein levels in cultured cells from FA patients. In a pilot clinical trial, 12 patients were treated with human recombinant erythropoietin (rHeEPO) to establish whether it could produce an increment in frataxin levels [188]. After 8 weeks of treatment, only two patients experienced a net increment in frataxin levels in peripheral blood lymphocytes, patients who also showed a reduction in markers of oxidative stress. However, it is not yet clear whether or not erythropoietin raises frataxin levels within affected neurons. Since chromatin condensation around the GAA repeat is thought to be responsible, to some extent, for the low levels of frataxin mRNA expression in FA, histone deacetylase (HDAC) inhibitors have also been investigated with the same encouraging results [189, 190].
FA is a good candidate for treatment with gene therapy as it is a monogenic disease and an increases in frataxin level could substantially improved the symptoms given that healthy carriers may have around 40%–50% of normal frataxin levels [191].
A human frataxin cDNA has been delivered into FA patient fibroblasts using lentiviral or adeno-associated viral (AAV) vectors, which has resulted in a partial correction of their sensitivity to oxidant stress [192]. However, the nonphysiological overexpression of frataxin driven by these vectors has been shown to be cytopathological.
Interestingly, the expression of frataxin cDNA to “physiological” levels driven by a HSV-1 amplicon vector can rescue the neurodegeneration triggered by frataxin deficiency both in cultured neurons and in vivo [193]. These results constitute the first “proof of principle” that neurological function can be recovered through a gene therapy approach aimed at correcting frataxin deficiency.
As mentioned above, it seems that optimal frataxin levels are required for correct cell function, since both deficiency and massive overexpression are associated with cell pathology. Thus, physiologically regulated expression of frataxin gene seems to be important for the gene therapy to succeed in FA. An interesting possibility is the use of the entire genomic locus of frataxin, since its large size may ensure the inclusion of all the regulatory elements required for proper gene expression.
Previous reports of transgenic mice indeed support the use of the frataxin genomic locus to correct frataxin deficiency in vivo. Thus, the entire FXN gene within a human YAC clone of 370 kb was reported to rescue the embryonic lethality of frataxin knock-out mice [19]. Likewise, a human BAC clone of 188 kb containing the entire genomic FXN locus has also been demonstrated to rescue the lethal phenotype of frataxin deficiency [16]. This indicates that the FRDA-BAC contains all the regulatory elements needed for the FRDA gene to be expressed in a physiological fashion.
A slightly smaller FRDA-BAC of 135 kb containing the entire 80 kb FXN genomic locus has been used to generate an HSV-1 amplicon (iBAC-FRDA) in order to test its ability to correct the FA phenotype [194]. This vector is capable of restoring physiological levels of frataxin in fibroblasts from FA patients and hence, of almost completely rescuing the cell phenotype of susceptibility to oxidative stress. Another key issue when using this kind of vector is the ability to infect cells that are hard to transfect with other techniques [195]. Once FRDA expression from the iBAC has been demonstrated, the iBAC might be used for in vivo delivery of the FRDA gene to FA-affected regions since delivery of HSV-1 amplicons to determined brain regions has already been shown [127]. Indeed, we have results indicating that HSV-1 amplicons containing the entire frataxin genomic locus produce persistent gene expression in the nervous system in vivo (Corona et al., manuscript in preparation). All these results support the promise of these vectors for gene therapy of FA.
In this regard, it is particularly interesting that herpes viral vectors are now in clinical trials for gene therapy of chronic pain. The results of this trial will provide the first clues about the safety and efficacy of herpes viral vectors as “gene-delivery vehicles” in the human nervous system [196].
5. Conclusions
Artificial chromosomes and minichromosome-like episomes based on BACs containing entire genomic loci are very promising tools for gene therapy of inherited diseases caused by recessive mutations, such as haemophilia or Friedreich's ataxia. The development of vehicles capable of accommodate whole genes emerges like a priority due to the importance of correct and accurate expression of most genes. Even if this is the key question, we should not forget that the way these molecules are delivered into target cells is as important as the former. So, improved development of delivery methods should be as well crucial for the success of gene therapy using these large DNA molecules. In this respect, amplicon vectors based on herpes viruses are currently a very useful tool regarding average BACs but still much work is needed in order to increase their DNA capacity or finding alternative methods for delivery.
Acknowledgments
the work on gene therapy in the authors lab is supported by Grants from the “Plan Nacional de Investigación en Biomedicina” (SAF 2006-12782-C03-02 and SAF2009-10757), the “Comunidad Autónoma de Madrid” (Neurodegmodels, Ref S-SAL-0202-2006) and the “Fundación Alicia Koplowitz”. The Biomedical Network Research Centre for Rare Diseases “Centro de Investigación Biomédica en Red sobre Enfermedades Raras” (CIBERER) is supported by the “Instituto de Salud Carlos III” (ISC III). The authors also want to acknowledge Dr. Mark Sefton for paper revision.
References
- 1.Dai Y, Schwarz EM, Gu D, Zhang W-W, Sarvetnick N, Verma IM. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(5):1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowenstein PR, Mandel RJ, Xiong W-D, Kroeger K, Castro MG. Immune responses to adenovirus and adeno-associated vectors used for gene therapy of brain diseases: the role of immunological synapses in understanding the cell biology of neuroimmune interactions. Current Gene Therapy. 2007;7(5):347–360. doi: 10.2174/156652307782151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochanek S, Clemens PR, Mitani K, Chen H-H, Chan S, Caskey CT. A new adenoviral vector: replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and β-galactosidase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(12):5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochanek S, Schiedner G, Volpers C. High-capacity ’gutless’ adenoviral vectors. Current Opinion in Molecular Therapeutics. 2001;3(5):454–463. [PubMed] [Google Scholar]
- 5.Lowenstein PR, Thomas CE, Umana P, et al. High-capacity, helper-dependent, "gutless" adenoviral vectors for gene transfer into brain. Methods in Enzymology. 2002;346:292–311. doi: 10.1016/s0076-6879(02)46062-4. [DOI] [PubMed] [Google Scholar]
- 6.Palù G, Parolin C, Takeuchi Y, Pizzato M. Progress with retroviral gene vectors. Reviews in Medical Virology. 2000;10(3):185–202. doi: 10.1002/(sici)1099-1654(200005/06)10:3<185::aid-rmv285>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Düllmann J, Schiedlmeier B, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296(5567):p. 497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 8.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nature Reviews Genetics. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 9.Check E. Regulators split on gene therapy as patient shows signs of cancer. Nature. 2002;419(6907):545–546. doi: 10.1038/419545a. [DOI] [PubMed] [Google Scholar]
- 10.Naldini L, Blömer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 11.Reiser J, Harmison G, Kluepfel-Stahl S, Brady RO, Karlsson S, Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(26):15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pannell D, Osborne CS, Yao S, et al. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO Journal. 2000;19(21):5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaenisch R. Transgenic animals. Science. 1988;240(4858):1468–1474. doi: 10.1126/science.3287623. [DOI] [PubMed] [Google Scholar]
- 14.Pittman DD, Alderman EM, Tomkinson KN, Wang JH, Giles AR, Kaufman RJ. Biochemical, immunological, and in vivo functional characterization of B- domain-deleted factor VIII. Blood. 1993;81(11):2925–2935. [PubMed] [Google Scholar]
- 15.Huxley C. Exploring gene function: use of yeast artificial chromosome transgenesis. Methods. 1998;14(2):199–210. doi: 10.1006/meth.1997.0578. [DOI] [PubMed] [Google Scholar]
- 16.Sarsero JP, Li L, Holloway TP, et al. Human BAC-mediated rescue of the Friedreich ataxia knockout mutation in transgenic mice. Mammalian Genome. 2004;15(5):370–382. doi: 10.1007/s00335-004-3019-3. [DOI] [PubMed] [Google Scholar]
- 17.Brem G, Besenfelder U, Aigner B, et al. YAC transgenesis in farm animals: rescue of albinism in rabbits. Molecular Reproduction and Development. 1996;44(1):56–62. doi: 10.1002/(SICI)1098-2795(199605)44:1<56::AID-MRD6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 18.Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Research. 2001;10(2):83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 19.Pook MA, Al-Mahdawi S, Carroll CJ, et al. Rescue of the Friedreich’s ataxia knockout mouse by human YAC transgenesis. Neurogenetics. 2001;3(4):185–193. doi: 10.1007/s100480100118. [DOI] [PubMed] [Google Scholar]
- 20.Moreira PN, Pozueta J, Pérez-Crespo M, Valdivieso F, Gutiérrez-Adán A, Montoliu L. Improving the generation of genomic-type transgenic mice by ICSI. Transgenic Research. 2007;16(2):163–168. doi: 10.1007/s11248-007-9075-1. [DOI] [PubMed] [Google Scholar]
- 21.Manson AL, Trezise AEO, MacVinish LJ, et al. Complementation of null CF mice with a human CFTR YAC transgene. EMBO Journal. 1997;16(14):4238–4249. doi: 10.1093/emboj/16.14.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgson JG, Smith DJ, McCutcheon K, et al. Human huntingtin derived from YAC transgenes compensates for loss of murine huntingtin by rescue of the embryonic lethal phenotype. Human Molecular Genetics. 1996;5(12):1875–1885. doi: 10.1093/hmg/5.12.1875. [DOI] [PubMed] [Google Scholar]
- 23.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nature Reviews Genetics. 2001;2(10):769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 24.Muyrers JPP, Zhang Y, Stewart AF. Techniques: recombinogenic engineering—new options for cloning and manipulating DNA. Trends in Biochemical Sciences. 2001;26(5):325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 25.Narayanan K, Williamson R, Zhang Y, Stewart AF, Ioannou PA. Efficient and precise engineering of a 200 kb β-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Therapy. 1999;6(3):442–447. doi: 10.1038/sj.gt.3300901. [DOI] [PubMed] [Google Scholar]
- 26.Poser I, Sarov M, Hutchins JRA, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nature Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choo KHA. Centromere DNA dynamics: latent centromeres and neocentrome formation. American Journal of Human Genetics. 1997;61(6):1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurnit DM, Maio JJ. Variable satellite DNA’s in the African green monkey Cercopithecus aethiops. Chromosoma. 1974;45(4):387–400. doi: 10.1007/BF00283385. [DOI] [PubMed] [Google Scholar]
- 29.Potter SS, Jones RS. Unusual domains of human alphoid satellite DNA with contiguous non-satellite sequences: sequence analysis of a junction region. Nucleic Acids Research. 1983;11(10):3137–3153. doi: 10.1093/nar/11.10.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg H, Singer M, Rosenberg M. Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978;200(4340):394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- 31.Thayer RE, Singer MF, McCutchan TF. Sequence relationships between single repeat units of highly reiterated African Green monkey DNA. Nucleic Acids Research. 1981;9(1):169–181. doi: 10.1093/nar/9.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willard HF, Wayne JS. Hierarchical order in chromosome-specific human alpha setellite DNA. Trends in Genetics. 1987;3(7):192–198. [Google Scholar]
- 33.Larin Z, Fricker MD, Tyler-Smith C. De novo formation of several features of a centromere following introduction of a Y alphoid YAC into mammalian cells. Human Molecular Genetics. 1994;3(5):689–695. doi: 10.1093/hmg/3.5.689. [DOI] [PubMed] [Google Scholar]
- 34.Wevrick R, Willard HF. Long-range organization of tandem arrays of α satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(23):9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nature Genetics. 1997;15(4):345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 36.Mejía JE, Alazami A, Willmott A, et al. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics. 2002;79(3):297–304. doi: 10.1006/geno.2002.6704. [DOI] [PubMed] [Google Scholar]
- 37.Ohzeki J-I, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. Journal of Cell Biology. 2002;159(5):765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masumoto H, Ikeno M, Nakano M, et al. Assay of centromere function using a human artificial chromosome. Chromosoma. 1998;107(6-7):406–416. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- 39.Choo KHA. Centromerization. Trends in Cell Biology. 2000;10(5):182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- 40.du Sart D, Cancilla MR, Earle E, et al. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nature Genetics. 1997;16(2):144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 41.Depinet TW, Zackowski JL, Earnshaw WC, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Human Molecular Genetics. 1997;6(8):1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 42.Earnshaw WC, Ratrie H, III, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98(1):1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 43.Willard HF. Neocentromeres and human artificial chromosomes: an unnatural act. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(10):5374–5376. doi: 10.1073/pnas.111167398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JW, Pendon C, Yang J, Haywood N, Chand A, Brown WRA. Human mini-chromosomes with minimal centromeres. Human Molecular Genetics. 2000;9(12):1891–1902. doi: 10.1093/hmg/9.12.1891. [DOI] [PubMed] [Google Scholar]
- 45.Lo AWI, Liao GC-C, Rocchi M, Choo KHA. Extreme reduction of chromosome-specific α-satellite array is unusually common in human chromosome 21. Genome Research. 1999;9(10):895–908. doi: 10.1101/gr.9.10.895. [DOI] [PubMed] [Google Scholar]
- 46.Ebersole TA, Ross A, Clark E, et al. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Human Molecular Genetics. 2000;9(11):1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 47.Ikeno M, Grimes B, Okazaki T, et al. Construction of YAC-based mammalian artificial chromosomes. Nature Biotechnology. 1998;16(5):431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 48.Kotzamanis G, Cheung W, Abdulrazzak H, et al. Construction of human artificial chromosome vectors by recombineering. Gene. 2005;351:29–38. doi: 10.1016/j.gene.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Willard HF. Chromosome manipulation: a systematic approach toward understanding human chromosome structure and function. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(14):6847–6850. doi: 10.1073/pnas.93.14.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke DT, Carle GF, Olson MV. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 51.Jakobovits A, Moore AL, Green LL, et al. Germ-line transmission and expression of a human-derived yeast artificial chromosome. Nature. 1993;362(6417):255–258. doi: 10.1038/362255a0. [DOI] [PubMed] [Google Scholar]
- 52.Alazami AM, Mejía JE, Monaco ZL. Human artificial chromosomes containing chromosome 17 alphoid DNA maintain an active centromere in murine cells but are not stable. Genomics. 2004;83(5):844–851. doi: 10.1016/j.ygeno.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Grimes BR, Schindelhauer D, McGill NI, Ross A, Ebersole TA, Cooke HJ. Stable gene expression from a mammalian artificial chromosome. EMBO Reports. 2001;2(10):910–914. doi: 10.1093/embo-reports/kve187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mejía JE, Willmott A, Levy E, Earnshaw WC, Larin Z. Functional complementation of a genetic deficiency with human artificial chromosomes. American Journal of Human Genetics. 2001;69(2):315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mejía JE, Larin Z. The assembly of large BACs by in vivo recombination. Genomics. 2000;70(2):165–170. doi: 10.1006/geno.2000.6372. [DOI] [PubMed] [Google Scholar]
- 56.Kakeda M, Hiratsuka M, Nagata K, et al. Human artificial chromosome (HAC) vector provides long-term therapeutic transgene expression in normal human primary fibroblasts. Gene Therapy. 2005;12(10):852–856. doi: 10.1038/sj.gt.3302483. [DOI] [PubMed] [Google Scholar]
- 57.Killary AM, Fournier REK. Microcell fusion. Methods in Enzymology. 1995;254:133–152. doi: 10.1016/0076-6879(95)54011-3. [DOI] [PubMed] [Google Scholar]
- 58.Hoshiya H, Kazuki Y, Abe S, et al. A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Molecular Therapy. 2009;17(2):309–317. doi: 10.1038/mt.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kazuki Y, Hiratsuka M, Takiguchi M, et al. Complete genetic correction of iPS cells from Duchenne muscular dystrophy. Molecular Therapy. 2009;18(2):386–393. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudd MK, Mays RW, Schwartz S, Willard HF. Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Molecular and Cellular Biology. 2003;23(21):7689–7697. doi: 10.1128/MCB.23.21.7689-7697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spence JM, Mills W, Mann K, Huxley C, Farr CJ. Increased missegregation and chromosome loss with decreasing chromosome size in vertebrate cells. Chromosoma. 2006;115(1):60–74. doi: 10.1007/s00412-005-0032-6. [DOI] [PubMed] [Google Scholar]
- 62.Shen MH, Yang JW, Yang J, Pendon C, Brown WRA. The accuracy of segregation of human mini-chromosomes varies in different vertebrate cell lines, correlates with the extent of centromere formation and provides evidence for a trans-acting centromere maintenance activity. Chromosoma. 2000;109(8):524–535. doi: 10.1007/s004120000110. [DOI] [PubMed] [Google Scholar]
- 63.Sclimenti CR, Calos MP. Epstein-Barr virus vectors for gene expression and transfer. Current Opinion in Biotechnology. 1998;9(5):476–479. doi: 10.1016/s0958-1669(98)80032-3. [DOI] [PubMed] [Google Scholar]
- 64.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Molecular and Cellular Biology. 1985;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 67.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Molecular and Cellular Biology. 1985;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lupton S, Levine AJ. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Molecular and Cellular Biology. 1985;5(10):2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 70.Gahn TA, Schildkraut CL. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 71.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Molecular and Cellular Biology. 1986;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yates JL, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. Journal of Virology. 1991;65(1):483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haase SB, Calos MP. Replication control of autonomously replicating human sequences. Nucleic Acids Research. 1991;19(18):5053–5058. doi: 10.1093/nar/19.18.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krysan PJ, Calos MP. Epstein-Barr virus-based vectors that replicate in rodent cells. Gene. 1993;136(1-2):137–143. doi: 10.1016/0378-1119(93)90457-e. [DOI] [PubMed] [Google Scholar]
- 75.Nayyar VK, Shire K, Frappier L. Mitotic chromosome interactions of Epstein-Barr nuclear antigen 1 (EBNA1) and human EBNA1-bindingprotein 2 (EBP2) Journal of Cell Science. 2009;122(23):4341–4350. doi: 10.1242/jcs.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okayama H, Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Molecular and Cellular Biology. 1983;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Margolskee RF, Kavathas P, Berg P. Epstein-Barr virus shuttle vector for stable episomal replication of cDNA expression libraries in human cells. Molecular and Cellular Biology. 1988;8(7):2837–2847. doi: 10.1128/mcb.8.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banerjee S, Livanos E, Vos J-MH. Therapeutic gene delivery in human B-lymphoblastoid cells by engineered non-transforming infectious Epstein-Barr virus. Nature Medicine. 1995;1(12):1303–1308. doi: 10.1038/nm1295-1303. [DOI] [PubMed] [Google Scholar]
- 79.Lei DC, Kunzelmann K, Koslowsky T, et al. Episomal expression of wild-type CFTR corrects cAMP-dependent chloride transport in respiratory epithelial cells. Gene Therapy. 1996;3(5):427–436. [PubMed] [Google Scholar]
- 80.Mücke S, Polack A, Pawlita M, et al. Suitability of Epstein-Barr virus-based episomal vectors for expression of cytokine genes in human lymphoma cells. Gene Therapy. 1997;4(2):82–92. doi: 10.1038/sj.gt.3300363. [DOI] [PubMed] [Google Scholar]
- 81.Young JM, Cheadle C, Foulke JS, Jr., Drohan WN, Sarver N. Utilization of an Epstein-Barr virus replicon as a eukaryotic expression vector. Gene. 1988;62(2):171–185. doi: 10.1016/0378-1119(88)90556-2. [DOI] [PubMed] [Google Scholar]
- 82.Cachianes G, Ho C, Weber RF, Williams SR, Goeddel DV, Leung DW. Epstein-Barr virus-derived vectors for transient and stable expression of recombinant proteins. BioTechniques. 1993;15(2):255–258. [PubMed] [Google Scholar]
- 83.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Molecular and Cellular Biology. 1996;16(9):5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harris A, Young BD, Griffin BE. Random associated of Epstein-Barr virus genomes with host cell metaphase chromosome in Burkitt’s lymphoma-derived cell lines. Journal of Virology. 1985;56(1):328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huertas D, Howe S, McGuigan A, Huxley C. Expression of the human CFTR gene from episomal oriP-EBNA1-YACs in mouse cells. Human Molecular Genetics. 2000;9(4):617–629. doi: 10.1093/hmg/9.4.617. [DOI] [PubMed] [Google Scholar]
- 86.Kaname T, Huxley C. Simple and efficient vectors for retrofitting BACs and PACs with mammalian neoR and EGFP marker genes. Gene. 2001;266(1-2):147–153. doi: 10.1016/s0378-1119(01)00375-4. [DOI] [PubMed] [Google Scholar]
- 87.Magin-Lachmann C, Kotzamanis G, D’Aiuto L, Cooke H, Huxley C, Wagner E. In vitro and in vivo delivery of intact BAC DNA—comparison of different methods. Journal of Gene Medicine. 2004;6(2):195–209. doi: 10.1002/jgm.481. [DOI] [PubMed] [Google Scholar]
- 88.Magin-Lachman C, Kotzamanis G, D’Aiuto L, Wagner E, Huxley C. Retrofitting BACs with G418 resistance, luciferase, and oriP and EBNA-1—new vectors for in vitro and in vivo delivery. BMC Biotechnology. 2003;3, article no. 2 doi: 10.1186/1472-6750-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Courvalin P, Goussard S, Grillot-Courvalin C. Gene transfer from bacteria to mammalian cells. Comptes Rendus de l’Academie des Sciences. Serie III. 1995;318(12):1207–1212. [PubMed] [Google Scholar]
- 90.Grillot-Courvalin C, Goussard S, Courvalin P. Wild-type intracellular bacteria deliver DNA into mammalian cells. Cellular Microbiology. 2002;4(3):177–186. doi: 10.1046/j.1462-5822.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- 91.Schaffner W. Direct transfer of cloned genes from bacteria to mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(4 I):2163–2167. doi: 10.1073/pnas.77.4.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grillot-Courvalin C, Goussard S, Courvalin P. Bacteria as gene delivery vectors for mammalian cells. Current Opinion in Biotechnology. 1999;10(5):477–481. doi: 10.1016/s0958-1669(99)00013-0. [DOI] [PubMed] [Google Scholar]
- 93.Weiss S, Chakraborty T. Transfer of eukaryotic expression plasmids to mammalian host cells by bacterial carriers. Current Opinion in Biotechnology. 2001;12(5):467–472. doi: 10.1016/s0958-1669(00)00247-0. [DOI] [PubMed] [Google Scholar]
- 94.Xu DQ, Zhang L, Kopecko DJ, et al. Bacterial delivery of siRNAs: a new approach to solid tumor therapy. Methods in Molecular Biology. 2009;487:161–187. doi: 10.1007/978-1-60327-547-7_8. [DOI] [PubMed] [Google Scholar]
- 95.Chamekh M, Phalipon A, Quertainmont R, Salmon I, Sansonetti P, Allaoui A. Delivery of biologically active anti-inflammatory cytokines IL-10 and IL-1ra in vivo by the Shigella type III secretion apparatus. Journal of Immunology. 2008;180(6):4292–4298. doi: 10.4049/jimmunol.180.6.4292. [DOI] [PubMed] [Google Scholar]
- 96.Stritzker J, Pilgrim S, Szalay AA, Goebel W. Prodrug converting enzyme gene delivery by L. monocytogenes. BMC Cancer. 2008;8, article no. 94 doi: 10.1186/1471-2407-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Al-Mariri A, Tibor A, Lestrate P, Mertens P, De Bolle X, Letesson J-J. Yersinia enterocolitica as a vehicle for a naked DNA vaccine encoding Brucella abortus bacterioferritin or P39 antigen. Infection and Immunity. 2002;70(4):1915–1923. doi: 10.1128/IAI.70.4.1915-1923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sasaki T, Fujimori M, Hamaji Y, et al. Genetically engineered Bifidobacterium longum for tumor-targeting enzyme-prodrug therapy of autochthonous mammary tumors in rats. Cancer Science. 2006;97(7):649–657. doi: 10.1111/j.1349-7006.2006.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Michl P, Gress TM. Bacteria and bacterial toxins as therapeutic agents for solid tumors. Current Cancer Drug Targets. 2004;4(8):689–702. doi: 10.2174/1568009043332727. [DOI] [PubMed] [Google Scholar]
- 100.Lee C-H, Wu C-L, Shiau A-L. Systemic administration of attenuated Salmonella choleraesuis carrying thrombospondin-1 gene leads to tumor-specific transgene expression, delayed tumor growth and prolonged survival in the murine melanoma model. Cancer Gene Therapy. 2005;12(2):175–184. doi: 10.1038/sj.cgt.7700777. [DOI] [PubMed] [Google Scholar]
- 101.Ryan RM, Green J, Williams PJ, et al. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Therapy. 2009;16(3):329–339. doi: 10.1038/gt.2008.188. [DOI] [PubMed] [Google Scholar]
- 102.Li Z, Fallon J, Mandeli J, Wetmur J, Woo SLC. A genetically enhanced anaerobic bacterium for oncopathic therapy of pancreatic cancer. Journal of the National Cancer Institute. 2008;100(19):1389–1400. doi: 10.1093/jnci/djn308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Theys J, Landuyt W, Nuyts S, et al. Specific targeting of cytosine deaminase to solid tumors by engineered Clostridium acetobutylicum. Cancer Gene Therapy. 2001;8(4):294–297. doi: 10.1038/sj.cgt.7700303. [DOI] [PubMed] [Google Scholar]
- 104.Ning J-F, Zhu W, Xu J-P, Zheng C-Y, Meng X-L. Oral delivery of DNA vaccine encoding VP28 against white spot syndrome virus in crayfish by attenuated Salmonella typhimurium. Vaccine. 2009;27(7):1127–1135. doi: 10.1016/j.vaccine.2008.11.075. [DOI] [PubMed] [Google Scholar]
- 105.Abdul-Wahid A, Faubert G. Mucosal delivery of a transmission-blocking DNA vaccine encoding Giardia lamblia CWP2 by Salmonella typhimurium bactofection vehicle. Vaccine. 2007;25(50):8372–8383. doi: 10.1016/j.vaccine.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 106.Sandvig K, van Deurs B. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Therapy. 2005;12(11):865–872. doi: 10.1038/sj.gt.3302525. [DOI] [PubMed] [Google Scholar]
- 107.Grillot-Courvalin C, Goussard S, Huetz F, Ojcius DM, Courvalin P. Functional gene transfer from intracellular bacteria to mammalian cells. Nature Biotechnology. 1998;16(9):862–866. doi: 10.1038/nbt0998-862. [DOI] [PubMed] [Google Scholar]
- 108.Castagliuolo I, Beggiao E, Brun P, et al. Engineered E. coli delivers therapeutic genes to the colonic mucosa. Gene Therapy. 2005;12(13):1070–1078. doi: 10.1038/sj.gt.3302493. [DOI] [PubMed] [Google Scholar]
- 109.Isberg RR, Voorhis DL, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50(5):769–778. doi: 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- 110.Leong JM, Fournier RS, Isberg RR. Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO Journal. 1990;9(6):1979–1989. doi: 10.1002/j.1460-2075.1990.tb08326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leong JM, Morrissey PE, Isberg RR. A 76-amino acid disulfide loop in the Yersinia pseudotuberculosis invasin protein is required for integrin receptor recognition. Journal of Biological Chemistry. 1993;268(27):20524–20532. [PubMed] [Google Scholar]
- 112.Isberg RR, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317(6034):262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 113.Alouf JE. Cholesterol-binding cytolytic protein toxins. International Journal of Medical Microbiology. 2000;290(4-5):351–356. doi: 10.1016/S1438-4221(00)80039-9. [DOI] [PubMed] [Google Scholar]
- 114.Geoffroy C, Gaillard J-L, Alouf JE, Berche P. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infection and Immunity. 1987;55(7):1641–1646. doi: 10.1128/iai.55.7.1641-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cossart P, Vicente MF, Mengaud J, Baquero F, Perez-Diaz JC, Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infection and Immunity. 1989;57(11):3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Laner A, Goussard S, Ramalho AS, et al. Bacterial transfer of large functional genomic DNA into human cells. Gene Therapy. 2005;12(21):1559–1572. doi: 10.1038/sj.gt.3302576. [DOI] [PubMed] [Google Scholar]
- 117.Narayanan K, Warburton PE. DNA modification and functional delivery into human cells using Escherichia coli DH10B. Nucleic Acids Research. 2003;31(9, article no. e51) doi: 10.1093/nar/gng051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pérez-Luz S, Abdulrazzak H, Grillot-Courvalin C, Huxley C. Factor VIII mRNA expression from a BAC carrying the intact locus made by homologous recombination. Genomics. 2007;90(5):610–619. doi: 10.1016/j.ygeno.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 119.Poffenberger KL, Tabares E, Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proceedings of the National Academy of Sciences of the United States of America. 1983;80(9 I):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oehmig A, Fraefel C, Breakefield XO, Ackermann M. Herpes simplex virus type 1 amplicons and their hybrid virus partners, EBV, AAV, and retrovirus. Current Gene Therapy. 2004;4(4):385–408. doi: 10.2174/1566523043346129. [DOI] [PubMed] [Google Scholar]
- 121.Wade-Martins R, Saeki Y, Chiocca EA. Infectious delivery of a 135-kb LDLR genomic locus leads to regulated complementation of low-density lipoprotein receptor deficiency in human cells. Molecular Therapy. 2003;7(5):604–612. doi: 10.1016/s1525-0016(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 122.Müller L, Saydam O, Saeki Y, Heid I, Fraefel C. Gene transfer into hepatocytes mediated by herpes simplex virus-Epstein-Barr virus hybrid amplicons. Journal of Virological Methods. 2005;123(1):65–72. doi: 10.1016/j.jviromet.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 123.Epstein AL, Marconi P, Argnani R, Manservigi R. HSV-1-derived recombinant and amplicon vectors for gene transfer and gene therapy. Current Gene Therapy. 2005;5(5):445–458. doi: 10.2174/156652305774329285. [DOI] [PubMed] [Google Scholar]
- 124.Johnson PA, Miyanohara A, Levine F, Cahill T, Friedmann T. Cytotoxicity of a replication-defective mutant of herpes simplex virus type I. Journal of Virology. 1992;66(5):2952–2965. doi: 10.1128/jvi.66.5.2952-2965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saeki Y, Fraefel C, Ichikawa T, Breakefield XO, Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Molecular Therapy. 2001;3(4):591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- 126.Saeki Y, Ichikawa T, Saeki A, et al. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Human Gene Therapy. 1998;9(18):2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 127.Agudo M, Luis Trejo J, Lim F, et al. Highly efficient and specific gene transfer to purkinje cells in vivo using a herpes simplex virus I amplicon. Human Gene Therapy. 2002;13(5):665–674. doi: 10.1089/10430340252837251. [DOI] [PubMed] [Google Scholar]
- 128.Berges BK, Wolfe JH, Fraser NW. Transduction of brain by herpes simplex virus vectors. Molecular Therapy. 2007;15(1):20–29. doi: 10.1038/sj.mt.6300018. [DOI] [PubMed] [Google Scholar]
- 129.Fink DJ, DeLuca NA, Goins WF, Glorioso JC. Gene transfer to neurons using herpes simplex virus-based vectors. Annual Review of Neuroscience. 1996;19:265–287. doi: 10.1146/annurev.ne.19.030196.001405. [DOI] [PubMed] [Google Scholar]
- 130.Frampton AR, Jr., Goins WF, Nakano K, Burton EA, Glorioso JC. HSV trafficking and development of gene therapy vectors with applications in the nervous system. Gene Therapy. 2005;12(11):891–901. doi: 10.1038/sj.gt.3302545. [DOI] [PubMed] [Google Scholar]
- 131.Oehmig A, Fraefel C, Breakefield XO. Update on herpesvirus amplicon vectors. Molecular Therapy. 2004;10(4):630–643. doi: 10.1016/j.ymthe.2004.06.641. [DOI] [PubMed] [Google Scholar]
- 132.Jin BK, Belloni M, Conti B, et al. Prolonged in vivo gene expression driven by a tyrosine hydroxylase promoter in a defective herpes simplex virus amplicon vector. Human Gene Therapy. 1996;7(16):2015–2024. doi: 10.1089/hum.1996.7.16-2015. [DOI] [PubMed] [Google Scholar]
- 133.Moralli D, Simpson KM, Wade-Martins R, Monaco ZL. A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Reports. 2006;7(9):911–918. doi: 10.1038/sj.embor.7400768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Borst EM, Messerle M. Construction of a cytomegalovirus-based amplicon: a vector with a unique transfer capacity. Human Gene Therapy. 2003;14(10):959–970. doi: 10.1089/104303403766682223. [DOI] [PubMed] [Google Scholar]
- 135.Mannucci PM, Tuddenham EGD. The hemophilias—from royal genes to gene therapy. New England Journal of Medicine. 2001;344(23):1773–1779. doi: 10.1056/NEJM200106073442307. [DOI] [PubMed] [Google Scholar]
- 136.Chao H, Samulski RJ, Bellinger DA, Monahan PE, Nichols TC, Walsh CE. Persistent expression of canine factor IX in hemophilia B canines. Gene Therapy. 1999;6(10):1695–1704. doi: 10.1038/sj.gt.3301024. [DOI] [PubMed] [Google Scholar]
- 137.Cherington V, et al. Retroviral vector-modified bone marrow stromal cells secrete biologically active factor IX in vitro and transiently deliver therapeutic levels of human factor IX to the plasma of dogs after reinfusion. Human Gene Therapy. 1998;9(10):1397–1407. doi: 10.1089/hum.1998.9.10-1397. [DOI] [PubMed] [Google Scholar]
- 138.Fang B, Eisensmith RC, Wang H, et al. Gene therapy for hemophilia B: host immunosuppression prolongs the therapeutic effect of adenovirus-mediated factor IX expression. Human Gene Therapy. 1995;6(8):1039–1044. doi: 10.1089/hum.1995.6.8-1039. [DOI] [PubMed] [Google Scholar]
- 139.Niemeyer GP, Herzog RW, Mount J, et al. Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy. Blood. 2009;113(4):797–806. doi: 10.1182/blood-2008-10-181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kay MA, High K. Gene therapy for the hemophilias. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):9973–9975. doi: 10.1073/pnas.96.18.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Connelly S, Smith TAG, Dhir G, et al. In vivo gene delivery and expression of physiological levels of functional human factor VIII in mice. Human Gene Therapy. 1995;6(2):185–193. doi: 10.1089/hum.1995.6.2-185. [DOI] [PubMed] [Google Scholar]
- 142.Chuah MKL, Vandendriessche T, Morgan RA. Development and analysis of retroviral vectors expressing human factor VIII as a potential gene therapy for hemophilia A. Human Gene Therapy. 1995;6(11):1363–1377. doi: 10.1089/hum.1995.6.11-1363. [DOI] [PubMed] [Google Scholar]
- 143.Eaton DL, Hass PE, Riddle L. Characterization of recombinant human factor VIII. Journal of Biological Chemistry. 1987;262(7):3285–3290. [PubMed] [Google Scholar]
- 144.Toole JJ, et al. A large region (approximately equal to 95 kDa) of human factor VIII is dispensable for in vitro procoagulant activity. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(16):5939–5942. doi: 10.1073/pnas.83.16.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Eaton DL, Wood WI, Eaton D, et al. Construction and characterization of an active factor VIII variant lacking the central one-third of the molecule. Biochemistry. 1986;25(26):8343–8347. doi: 10.1021/bi00374a001. [DOI] [PubMed] [Google Scholar]
- 146.Yang Y, Wilson JM. Clearance of adenovirus-infected hepatocytes by MHC class I-restricted CD4+ CTLs in vivo. Journal of Immunology. 1995;155(5):2564–2570. [PubMed] [Google Scholar]
- 147.Connelly S, Gardner JM, Lyons RM, McClelland A, Kaleko M. Sustained expression of therapeutic levels of human factor VIII in mice. Blood. 1996;87(11):4671–4677. [PubMed] [Google Scholar]
- 148.Balagué C, Zhou J, Dai Y, et al. Sustained high-level expression of full-length human factor VIII and restoration of clotting activity in hemophilic mice using a minimal adenovirus vector. Blood. 2000;95(3):820–828. [PubMed] [Google Scholar]
- 149.Connelly S, Mount J, Mauser A, et al. Complete short-term correction of canine hemophilia A by in vivo gene therapy. Blood. 1996;88(10):3846–3853. [PubMed] [Google Scholar]
- 150.Andrews JL, Shirley PS, Iverson WO, et al. Evaluation of the duration of human factor VIII expression in nonhuman primates after systemic delivery of an adenoviral vector. Human Gene Therapy. 2002;13(11):1331–1336. doi: 10.1089/104303402760128559. [DOI] [PubMed] [Google Scholar]
- 151.Brown BD, Shi CX, Powell S, Hurlbut D, Graham FL, Lillicrap D. Helper-dependent adenoviral vectors mediate therapeutic factor VIII expression for several months with minimal accompanying toxicity in a canine model of severe hemophilia A. Blood. 2004;103(3):804–810. doi: 10.1182/blood-2003-05-1426. [DOI] [PubMed] [Google Scholar]
- 152.Andrews JL, Kadan MJ, Gorziglia MI, Kaleko M, Connelly S. Generation and characterization of E1/E2a/E3/E4-deficient adenoviral vectors encoding human factor VIII. Molecular Therapy. 2001;3(3):329–336. doi: 10.1006/mthe.2001.0264. [DOI] [PubMed] [Google Scholar]
- 153.Hoeben RC, Van Der Jagt RCM, Schoute F, et al. Expression of functional factor VIII in primary human skin fibroblasts after retrovirus-mediated gene transfer. Journal of Biological Chemistry. 1990;265(13):7318–7323. [PubMed] [Google Scholar]
- 154.Israel DI, Kaufman RJ. Retroviral-mediated transfer and amplification of a functional human factor VIII gene. Blood. 1990;75(5):1074–1080. [PubMed] [Google Scholar]
- 155.Chuah MKL, Brems H, Vanslembrouck V, Collen D, VandenDriessche T. Bone marrow stromal cells as targets for gene therapy of hemophilia A. Human Gene Therapy. 1998;9(3):353–365. doi: 10.1089/hum.1998.9.3-353. [DOI] [PubMed] [Google Scholar]
- 156.Fallaux FJ, Hoeben RC, Cramer SJ, et al. The human clotting factor VIII cDNA contains an autonomously replicating sequence consensus- and matrix attachment region-like sequence that binds a nuclear factor, represses heterologous gene expression, and mediates the transcriptional effects of sodium butyrate. Molecular and Cellular Biology. 1996;16(8):4264–4272. doi: 10.1128/mcb.16.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hoeben RC, Fallaux FJ, Cramer SJ, et al. Expression of the blood-clotting factor-VIII cDNA is repressed by a transcriptional silencer located in its coding region. Blood. 1995;85(9):2447–2454. [PubMed] [Google Scholar]
- 158.Koeberl DD, Halbert CL, Krumm A, Miller AD. Sequences within the coding regions of clotting factor VIII and CFTR block transcriptional elongation. Human Gene Therapy. 1995;6(4):469–479. doi: 10.1089/hum.1995.6.4-469. [DOI] [PubMed] [Google Scholar]
- 159.Lynch CM, Israel DI, Kaufman RJ, Miller AD. Sequences in the coding region of clotting factor VIII act as dominant inhibitors of RNA accumulation and protein production. Human Gene Therapy. 1993;4(3):259–272. doi: 10.1089/hum.1993.4.3-259. [DOI] [PubMed] [Google Scholar]
- 160.Byun J, Kim S-H, Mook Kim J, et al. Analysis of the relative level of gene expression from different retroviral vectors used for gene therapy. Gene Therapy. 1996;3(9):780–788. [PubMed] [Google Scholar]
- 161.Dwarki VJ, Belloni P, Nijjar T, et al. Gene therapy for hemophilia A: production of therapeutic levels of human factor VIII in vivo in mice. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(4):1023–1027. doi: 10.1073/pnas.92.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.VandenDriessche T, Vanslembrouck V, Goovaerts I, et al. Long-term expression of human coagulation factor VIII and correction of hemophilia A after in vivo retroviral gene transfer in factor VIII-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10379–10384. doi: 10.1073/pnas.96.18.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stein CS, Kang Y, Sauter SL, et al. In vivo treatment of hemophilia A and mucopolysaccharidosis type VII using nonprimate lentiviral vectors. Molecular Therapy. 2001;3(6):850–856. doi: 10.1006/mthe.2001.0325. [DOI] [PubMed] [Google Scholar]
- 164.Kootstra NA, Matsumura R, Verma IM. Efficient production of human FVIII in hemophilic mice using lentiviral vectors. Molecular Therapy. 2003;7(5):623–631. doi: 10.1016/s1525-0016(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 165.Chao H, Sun L, Bruce A, Xiao X, Walsh CE. Expression of human factor VIII by splicing between dimerized AAV vectors. Molecular Therapy. 2002;5(6):716–722. doi: 10.1006/mthe.2002.0607. [DOI] [PubMed] [Google Scholar]
- 166.Scallan CD, Lillicrap D, Jiang H, et al. Sustained phenotypic correction of canine hemophilia A using an adeno-associated viral vector. Blood. 2003;102(6):2031–2037. doi: 10.1182/blood-2003-01-0292. [DOI] [PubMed] [Google Scholar]
- 167.Youjin S, Jun Y. The treatment of hemophilia A: from protein replacement to AAV-mediated gene therapy. Biotechnology Letters. 2009;31(3):321–328. doi: 10.1007/s10529-008-9869-0. [DOI] [PubMed] [Google Scholar]
- 168.Roth DA, Tawa NE, Jr., O’Brien JM, Treco DA, Selden RF. Nonviral transfer of the gene encoding coagulation factor viii in patients with severe hemophilia A. New England Journal of Medicine. 2001;344(23):1735–1742. doi: 10.1056/NEJM200106073442301. [DOI] [PubMed] [Google Scholar]
- 169.Powell JS, Ragni MV, White GC, II, et al. Phase 1 trial of FVIII gene transfer for severe hemophilia A using a retroviral construct administered by peripheral intravenous infusion. Blood. 2003;102(6):2038–2045. doi: 10.1182/blood-2003-01-0167. [DOI] [PubMed] [Google Scholar]
- 170.Chuah MKL, Collen D, VandenDriassche T. Clinical gene transfer studies for hemophilia A. Seminars in Thrombosis and Hemostasis. 2004;30(2):249–256. doi: 10.1055/s-2004-825638. [DOI] [PubMed] [Google Scholar]
- 171.Kotzamanis G, Huxley C. Recombining overlapping BACs into a single larger BAC. BMC Biotechnology. 2004;4, article no. 1 doi: 10.1186/1472-6750-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Campuzano V, Montermini L, Moltò MD, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(5254):1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 173.Sakamoto N, Chastain PD, Parniewski P, et al. Sticky DNA: self-association properties of long GAA · TTC repeats in R · R · Y triplex structures from Friedreich’s ataxia. Molecular Cell. 1999;3(4):465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 174.Patel PI, Isaya G. Friedreich ataxia: from GAA triplet-repeat expansion to frataxin deficiency. American Journal of Human Genetics. 2001;69(1):15–24. doi: 10.1086/321283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Campuzano V, Montermini L, Lutz Y, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Human Molecular Genetics. 1997;6(11):1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 176.Babcock M, De Silva D, Oaks R, et al. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276(5319):1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 177.Adamec J, Rusnak F, Owen WG, et al. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. American Journal of Human Genetics. 2000;67(3):549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stehling O, Elsässer H, Brückel B, Mühlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Human Molecular Genetics. 2004;13(23):3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 179.Ristow M, Pfister MF, Yee AJ, et al. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(22):12239–12243. doi: 10.1073/pnas.220403797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koutnikova H, Campuzano V, Foury F, Dollé P, Cazzalini O, Koenig M. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nature Genetics. 1997;16(4):345–351. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- 181.Cossée M, Puccio H, Gansmuller A, et al. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Human Molecular Genetics. 2000;9(8):1219–1226. doi: 10.1093/hmg/9.8.1219. [DOI] [PubMed] [Google Scholar]
- 182.Puccio H, Simon D, Cossée M, et al. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nature Genetics. 2001;27(2):181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 183.Simon D, Seznec H, Gansmuller A, et al. Friedreich ataxia mouse models with progressive cerebellar and sensory ataxia reveal autophagic neurodegeneration in dorsal root ganglia. Journal of Neuroscience. 2004;24(8):1987–1995. doi: 10.1523/JNEUROSCI.4549-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Miranda CJ, Santos MM, Ohshima K, et al. Frataxin knockin mouse. FEBS Letters. 2002;512(1–3):291–297. doi: 10.1016/s0014-5793(02)02251-2. [DOI] [PubMed] [Google Scholar]
- 185.Al-Mahdawi S, Pinto RM, Varshney D, et al. GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics. 2006;88(5):580–590. doi: 10.1016/j.ygeno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pandolfo M. Drug insight: antioxidant therapy in inherited ataxias. Nature Clinical Practice Neurology. 2008;4(2):86–96. doi: 10.1038/ncpneuro0704. [DOI] [PubMed] [Google Scholar]
- 187.Acquaviva F, Castaldo I, Filla A, et al. Recombinant human erythropoietin increases frataxin protein expression without increasing mRNA expression. Cerebellum. 2008;7(3):360–365. doi: 10.1007/s12311-008-0036-x. [DOI] [PubMed] [Google Scholar]
- 188.Boesch S, Sturm B, Hering S, Goldenberg H, Poewe W, Scheiber-Mojdehkar B. Friedreich’s ataxia: clinical pilot trial with recombinant human erythropoietin. Annals of Neurology. 2007;62(5):521–524. doi: 10.1002/ana.21177. [DOI] [PubMed] [Google Scholar]
- 189.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nature Chemical Biology. 2006;2(10):551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 190.Rai M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;3(4, article no. e1958) doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pianese L, Turano M, Lo Casale MS, et al. Real time PCR quantification of frataxin mRNA in the peripheral blood leucocytes of Friedreich ataxia patients and carriers. Journal of Neurology, Neurosurgery and Psychiatry. 2004;75(7):1061–1063. doi: 10.1136/jnnp.2003.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fleming J, Spinoulas A, Zheng M, et al. Partial correction of sensitivity to oxidant stress in Friedreich ataxia patient fibroblasts by frataxin-encoding adeno-associated virus and lentivirus vectors. Human Gene Therapy. 2005;16(8):947–956. doi: 10.1089/hum.2005.16.947. [DOI] [PubMed] [Google Scholar]
- 193.Lim F, Palomo GM, Mauritz C, et al. Functional recovery in a Friedreich’s ataxia mouse model by frataxin gene transfer using an HSV-1 amplicon vector. Molecular Therapy. 2007;15(6):1072–1078. doi: 10.1038/sj.mt.6300143. [DOI] [PubMed] [Google Scholar]
- 194.Gomez-Sebastian S, Gimenez-Cassina A, Diaz-Nido J, Lim F, Wade-Martins R. Infectious delivery and expression of a 135kb human FRDA genomic DNA locus complements friedreich’s ataxia deficiency in human cells. Molecular Therapy. 2007;15(2):248–254. doi: 10.1038/sj.mt.6300021. [DOI] [PubMed] [Google Scholar]
- 195.Wade-Martins R, Smith ER, Tyminski E, Chiocca EA, Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nature Biotechnology. 2001;19(11):1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- 196.Glorioso JC, Fink DJ. Herpes vector-mediated gene transfer in the treatment of chronic pain. Molecular Therapy. 2009;17(1):13–18. doi: 10.1038/mt.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]